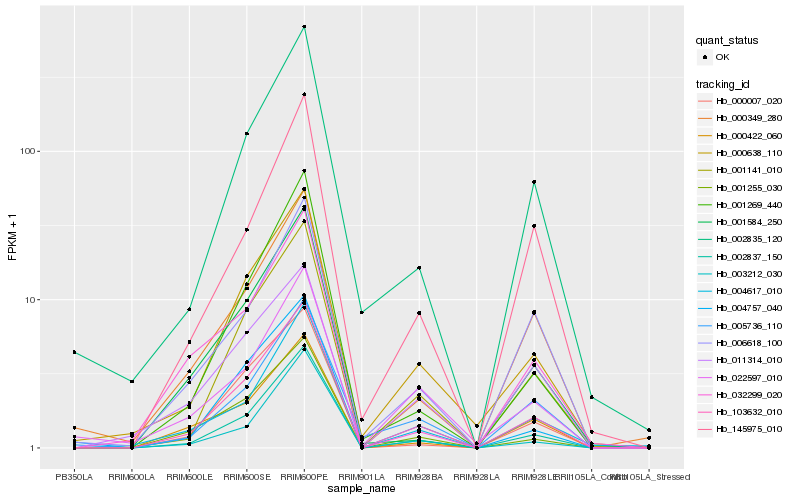
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103632_010 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_002837_150 |
0.0517196169 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 3 |
Hb_001141_010 |
0.0751239142 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 4 |
Hb_000638_110 |
0.0897212535 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 5 |
Hb_001255_030 |
0.0900224724 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840, partial [Sesamum indicum] |
| 6 |
Hb_004757_040 |
0.098616724 |
- |
- |
- |
| 7 |
Hb_000007_020 |
0.1107812399 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 8 |
Hb_004617_010 |
0.1124771655 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 9 |
Hb_002835_120 |
0.1137011639 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 10 |
Hb_000349_280 |
0.1139190362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000422_060 |
0.1148591812 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 12 |
Hb_001269_440 |
0.118313156 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 13 |
Hb_001584_250 |
0.1208914967 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 14 |
Hb_006618_100 |
0.1211792921 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 15 |
Hb_005736_110 |
0.1240501836 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 16 |
Hb_145975_010 |
0.1256784329 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_011314_010 |
0.1275775234 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_022597_010 |
0.131242062 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 19 |
Hb_003212_030 |
0.1315258967 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 20 |
Hb_032299_020 |
0.1326453723 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |