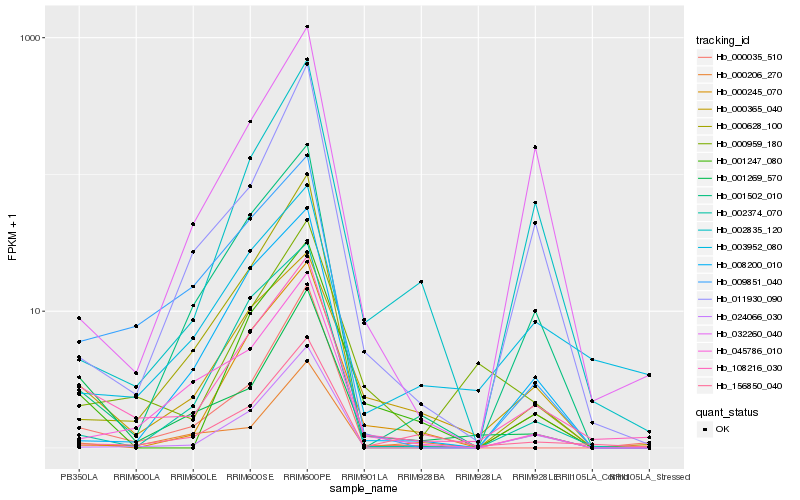
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108216_030 |
0.0 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000365_040 |
0.1482852533 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 3 |
Hb_003952_080 |
0.176101367 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_156850_040 |
0.1761317772 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 5 |
Hb_009851_040 |
0.1765058394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001269_570 |
0.1863035778 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 7 |
Hb_001247_080 |
0.1873621623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001502_010 |
0.1889545216 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 9 |
Hb_000035_510 |
0.1905329417 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 10 |
Hb_000959_180 |
0.1944143954 |
- |
- |
hypothetical protein JCGZ_09448 [Jatropha curcas] |
| 11 |
Hb_045786_010 |
0.1965348283 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_000628_100 |
0.1996728346 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 13 |
Hb_002835_120 |
0.2003795641 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 14 |
Hb_000245_070 |
0.2018691215 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 15 |
Hb_032260_040 |
0.2029273203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_024066_030 |
0.2031568235 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 17 |
Hb_011930_090 |
0.2034036953 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 18 |
Hb_008200_010 |
0.2035863282 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002374_070 |
0.2038673569 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000206_270 |
0.2045111835 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |