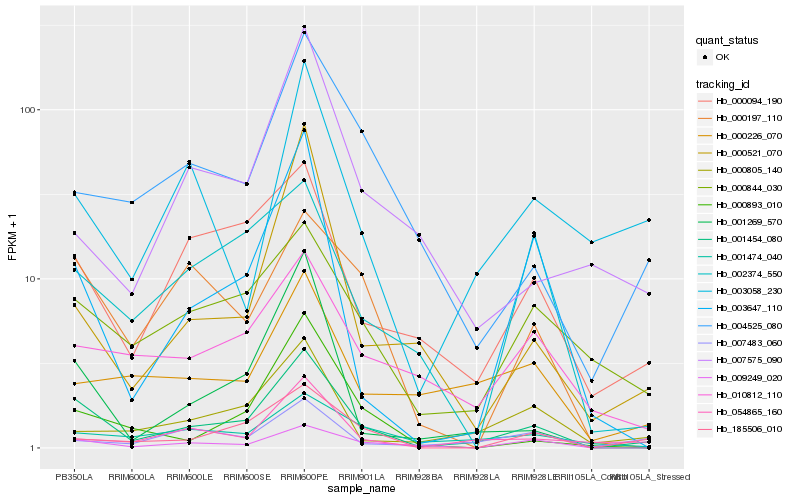
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_080 |
0.0 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 2 |
Hb_009249_020 |
0.2182837418 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 3 |
Hb_000094_190 |
0.2202658807 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 4 |
Hb_000844_030 |
0.2284192616 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 5 |
Hb_001269_570 |
0.2297787528 |
- |
- |
hypothetical protein POPTR_0001s03430g [Populus trichocarpa] |
| 6 |
Hb_003647_110 |
0.2370562252 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 7 |
Hb_185506_010 |
0.2401927809 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_000197_110 |
0.2443786554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_054865_160 |
0.2444757972 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 10 |
Hb_004525_080 |
0.245025876 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 11 |
Hb_010812_110 |
0.2499186962 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 12 |
Hb_001474_040 |
0.2502047755 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 13 |
Hb_000893_010 |
0.2554237835 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 14 |
Hb_000805_140 |
0.2558708594 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_000521_070 |
0.2588034369 |
- |
- |
PREDICTED: uncharacterized protein LOC105650323 [Jatropha curcas] |
| 16 |
Hb_000226_070 |
0.2615279827 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 17 |
Hb_007483_060 |
0.2668020045 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 18 |
Hb_003058_230 |
0.2668975999 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_002374_550 |
0.2697250655 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 20 |
Hb_007575_090 |
0.2732640695 |
- |
- |
putative. proteolipid subunit of vacuolar H+ ATPase, partial [Zea mays] |