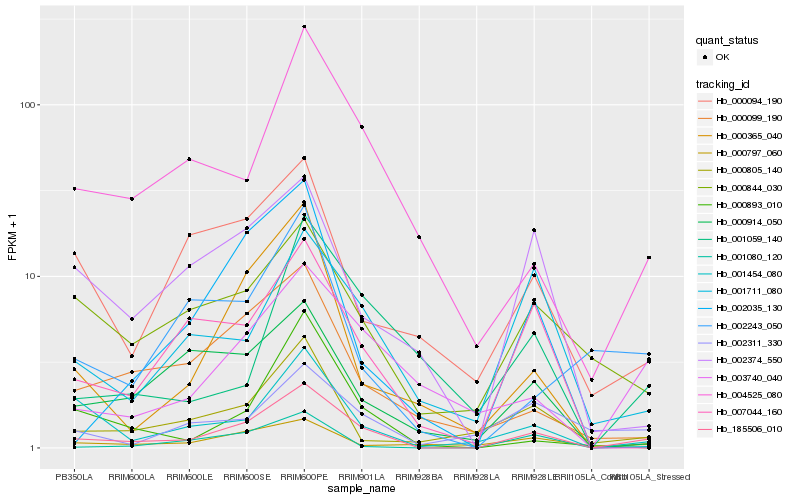
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185506_010 |
0.0 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_003740_040 |
0.2369698797 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase isozyme L3, putative [Ricinus communis] |
| 3 |
Hb_001454_080 |
0.2401927809 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_004525_080 |
0.241149428 |
- |
- |
Sugar transporter ERD6-like 6 [Morus notabilis] |
| 5 |
Hb_007044_160 |
0.246428097 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Populus euphratica] |
| 6 |
Hb_001080_120 |
0.2509625149 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 7 |
Hb_000099_190 |
0.2535686389 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 8 |
Hb_000844_030 |
0.2574684516 |
- |
- |
ent-kaurene oxidase, putative [Ricinus communis] |
| 9 |
Hb_000805_140 |
0.2594703946 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_001711_080 |
0.2621231005 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 11 |
Hb_000365_040 |
0.2636378239 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 12 |
Hb_000914_050 |
0.2670056445 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 13 |
Hb_002311_330 |
0.2704743172 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_002374_550 |
0.2738967422 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 15 |
Hb_000893_010 |
0.2739167847 |
- |
- |
PREDICTED: uncharacterized protein LOC105135422 [Populus euphratica] |
| 16 |
Hb_000094_190 |
0.2750663813 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 17 |
Hb_000797_060 |
0.2753334014 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 18 |
Hb_002035_130 |
0.2764697146 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 19 |
Hb_002243_050 |
0.2766815017 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_001059_140 |
0.2777490363 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |