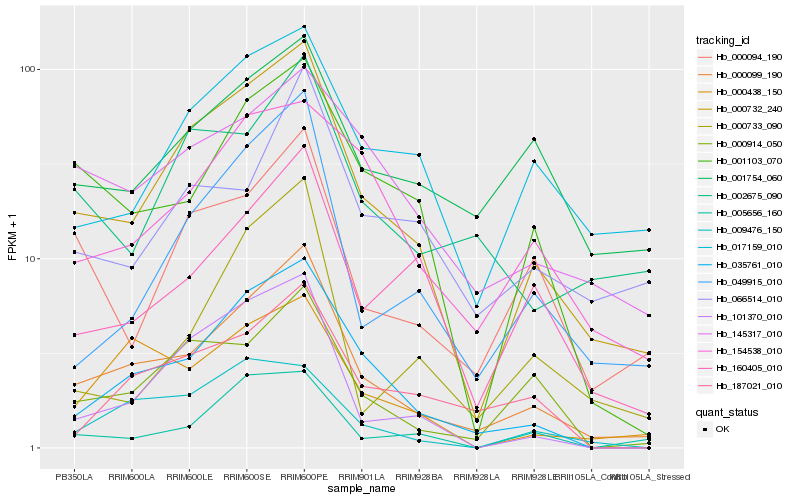
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_190 |
0.0 |
- |
- |
PREDICTED: putative cell division cycle ATPase [Jatropha curcas] |
| 2 |
Hb_000732_240 |
0.1120752763 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 3 |
Hb_035761_010 |
0.1334872869 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 4 |
Hb_049915_010 |
0.1733613244 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 5 |
Hb_000438_150 |
0.1757987766 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 6 |
Hb_000914_050 |
0.1761930174 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 7 |
Hb_001103_070 |
0.1762000915 |
- |
- |
Patellin-3, putative [Ricinus communis] |
| 8 |
Hb_187021_010 |
0.1866457876 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 9 |
Hb_160405_010 |
0.188411303 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 10 |
Hb_145317_010 |
0.1901131905 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 11 |
Hb_154538_010 |
0.1950645155 |
- |
- |
- |
| 12 |
Hb_000094_190 |
0.197431522 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 13 |
Hb_002675_090 |
0.1975482118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_101370_010 |
0.1981582655 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 15 |
Hb_001754_060 |
0.2009908496 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_066514_010 |
0.2029901216 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 17 |
Hb_017159_010 |
0.2037365967 |
- |
- |
- |
| 18 |
Hb_009476_150 |
0.2094547177 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 19 |
Hb_005656_160 |
0.2096737749 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 20 |
Hb_000733_090 |
0.2104205816 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |