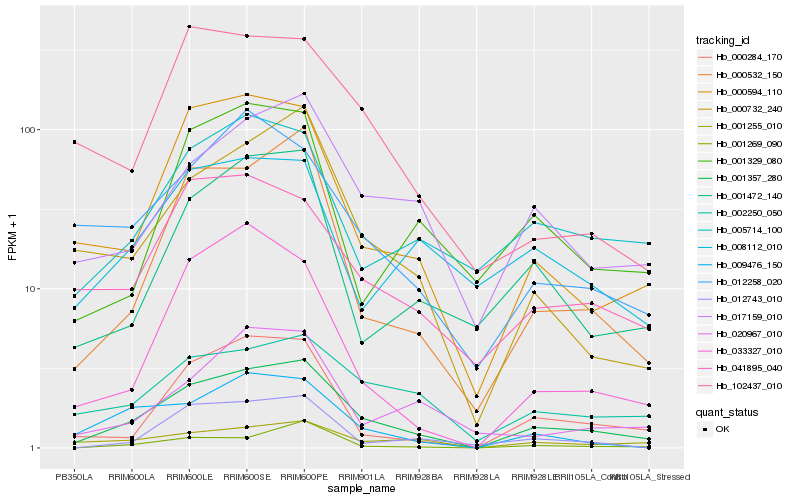
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_280 |
0.0 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 2 |
Hb_000532_150 |
0.1570187393 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 3 |
Hb_017159_010 |
0.1616032577 |
- |
- |
- |
| 4 |
Hb_000594_110 |
0.1646204768 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 5 |
Hb_000732_240 |
0.1683535814 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 6 |
Hb_000284_170 |
0.1709276594 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002250_050 |
0.1717433842 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 8 |
Hb_001255_010 |
0.1774967125 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 9 |
Hb_009476_150 |
0.1833800248 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 10 |
Hb_041895_040 |
0.183480127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001472_140 |
0.1843850025 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 12 |
Hb_005714_100 |
0.1880037707 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 13 |
Hb_001269_090 |
0.1924988272 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 14 |
Hb_012258_020 |
0.195102267 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_012743_010 |
0.197568227 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 16 |
Hb_001329_080 |
0.2005930341 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 17 |
Hb_020967_010 |
0.2017022987 |
- |
- |
- |
| 18 |
Hb_033327_010 |
0.2028748886 |
- |
- |
SYM8 [Pisum sativum] |
| 19 |
Hb_102437_010 |
0.2037531437 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_008112_010 |
0.2046506679 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |