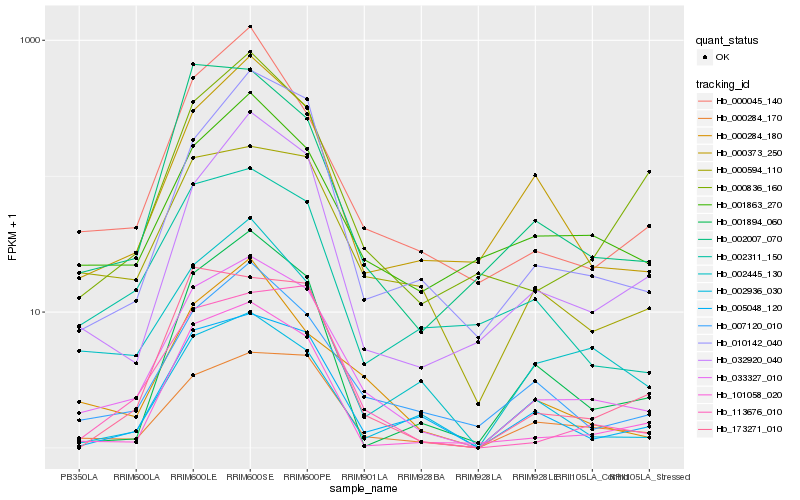
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033327_010 |
0.0 |
- |
- |
SYM8 [Pisum sativum] |
| 2 |
Hb_000284_170 |
0.1259964579 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_007120_010 |
0.1329444829 |
- |
- |
PREDICTED: uncharacterized protein LOC105634737 [Jatropha curcas] |
| 4 |
Hb_000594_110 |
0.1361277907 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 5 |
Hb_005048_120 |
0.1415224068 |
- |
- |
- |
| 6 |
Hb_000284_180 |
0.1511694821 |
- |
- |
hypothetical protein EUGRSUZ_B01865 [Eucalyptus grandis] |
| 7 |
Hb_010142_040 |
0.1527562345 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 8 |
Hb_000373_250 |
0.1553011303 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 9 |
Hb_101058_020 |
0.1559627606 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 10 |
Hb_002936_030 |
0.1597220385 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002445_130 |
0.160810286 |
- |
- |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 12 |
Hb_032920_040 |
0.1608751592 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 13 |
Hb_001863_270 |
0.1612198732 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 14 |
Hb_000836_160 |
0.1624326111 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 15 |
Hb_000045_140 |
0.16306731 |
- |
- |
PREDICTED: nematode resistance protein-like HSPRO2 [Jatropha curcas] |
| 16 |
Hb_113676_010 |
0.1631581691 |
- |
- |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 17 |
Hb_002311_150 |
0.1654410345 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 18 |
Hb_002007_070 |
0.169709537 |
- |
- |
PREDICTED: serrate RNA effector molecule homolog [Jatropha curcas] |
| 19 |
Hb_173271_010 |
0.1697484207 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 20 |
Hb_001894_060 |
0.1712220778 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |