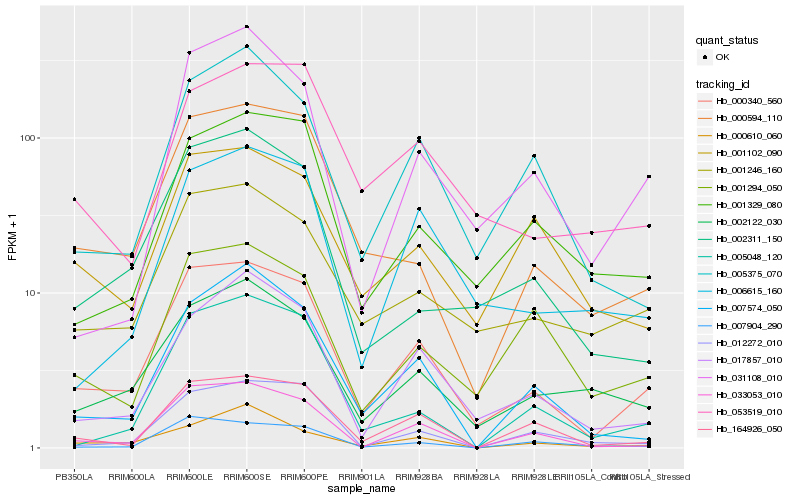
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_560 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_033053_010 |
0.1374826344 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 3 |
Hb_000594_110 |
0.1375959955 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 4 |
Hb_001246_160 |
0.1411632462 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 5 |
Hb_017857_010 |
0.1438921488 |
- |
- |
- |
| 6 |
Hb_164926_050 |
0.1450999384 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 7 |
Hb_002122_030 |
0.1473901734 |
- |
- |
- |
| 8 |
Hb_007574_050 |
0.1585885407 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 9 |
Hb_001329_080 |
0.1586481218 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 10 |
Hb_006615_160 |
0.158741384 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 11 |
Hb_002311_150 |
0.1590191814 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_005375_070 |
0.1609097256 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 13 |
Hb_053519_010 |
0.1667844083 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 14 |
Hb_001294_050 |
0.167762421 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 15 |
Hb_012272_010 |
0.1695510828 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_031108_010 |
0.1699334879 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 17 |
Hb_007904_290 |
0.1715362053 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 18 |
Hb_001102_090 |
0.1732540921 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 19 |
Hb_005048_120 |
0.1739701157 |
- |
- |
- |
| 20 |
Hb_000610_060 |
0.1742415204 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |