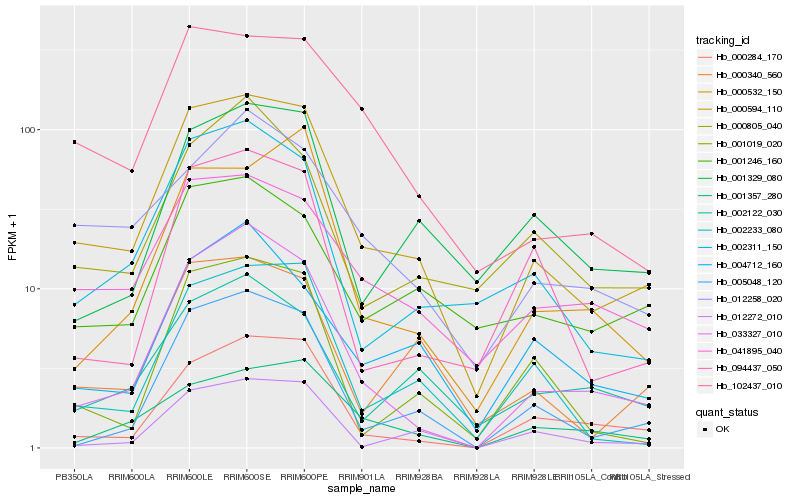
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_110 |
0.0 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 2 |
Hb_041895_040 |
0.1353900441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_102437_010 |
0.1360096017 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_033327_010 |
0.1361277907 |
- |
- |
SYM8 [Pisum sativum] |
| 5 |
Hb_000340_560 |
0.1375959955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002311_150 |
0.1405005895 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 7 |
Hb_002233_080 |
0.1444606973 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 8 |
Hb_000284_170 |
0.1505110123 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_005048_120 |
0.1528986798 |
- |
- |
- |
| 10 |
Hb_012258_020 |
0.1602774305 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_001246_160 |
0.163259899 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 12 |
Hb_001329_080 |
0.1642077947 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 13 |
Hb_002122_030 |
0.1643703118 |
- |
- |
- |
| 14 |
Hb_001357_280 |
0.1646204768 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 15 |
Hb_001019_020 |
0.1664382602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_094437_050 |
0.1664956599 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012272_010 |
0.1678526635 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_000805_040 |
0.1692147539 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 19 |
Hb_000532_150 |
0.1699088384 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 20 |
Hb_004712_160 |
0.1703391538 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |