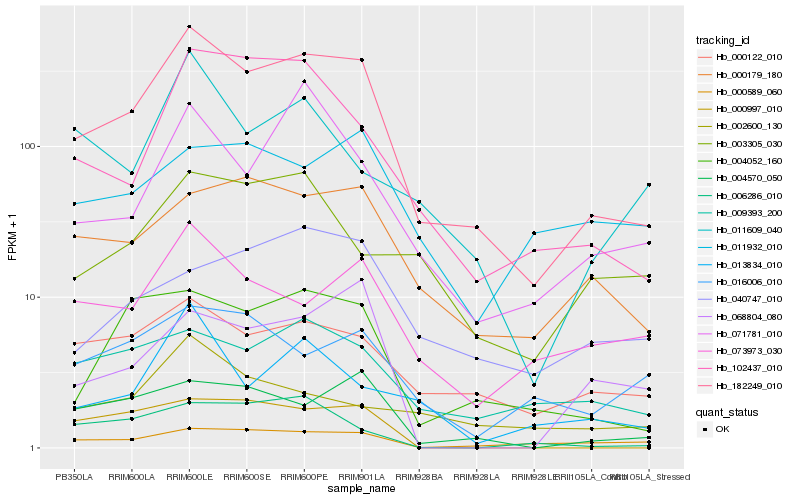
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182249_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004052_160 |
0.1734128628 |
- |
- |
Chain A, Rna Binding Protein |
| 3 |
Hb_000122_010 |
0.1793631529 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_000179_180 |
0.179805722 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 5 |
Hb_102437_010 |
0.1800832314 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004570_050 |
0.1839541731 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_009393_200 |
0.1901994241 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 8 |
Hb_000589_060 |
0.1973974085 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 9 |
Hb_073973_030 |
0.2007585345 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 10 |
Hb_071781_010 |
0.203789645 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |
| 11 |
Hb_013834_010 |
0.2178881878 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 12 |
Hb_040747_010 |
0.2184035334 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 13 |
Hb_016006_010 |
0.2197927677 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_003305_030 |
0.222989858 |
- |
- |
- |
| 15 |
Hb_011609_040 |
0.2232372862 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 40 [Jatropha curcas] |
| 16 |
Hb_068804_080 |
0.2272010127 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 17 |
Hb_002600_130 |
0.2286726111 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 18 |
Hb_000997_010 |
0.2288686509 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006286_010 |
0.2295837038 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 20 |
Hb_011932_010 |
0.2320503213 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |