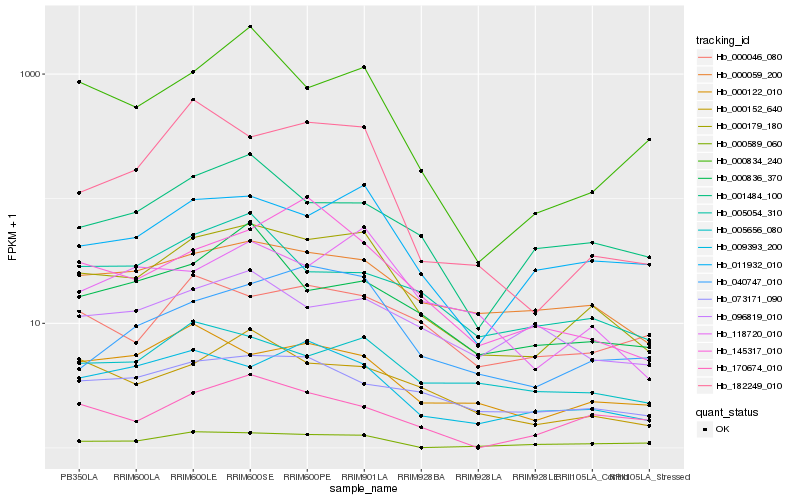
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_180 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 2 |
Hb_011932_010 |
0.1268311752 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 3 |
Hb_000059_200 |
0.1424423537 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 4 |
Hb_118720_010 |
0.1494271685 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 5 |
Hb_000152_640 |
0.1499593676 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 6 |
Hb_009393_200 |
0.1562532017 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 7 |
Hb_001484_100 |
0.1586281236 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 8 |
Hb_040747_010 |
0.1589498314 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 9 |
Hb_000589_060 |
0.1599240163 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 10 |
Hb_005656_080 |
0.1608178483 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 11 |
Hb_000122_010 |
0.1658195341 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_073171_090 |
0.1659357191 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 13 |
Hb_005054_310 |
0.170583718 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 14 |
Hb_000834_240 |
0.1707077286 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 15 |
Hb_000836_370 |
0.1746728384 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_145317_010 |
0.1789381333 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 17 |
Hb_182249_010 |
0.179805722 |
- |
- |
- |
| 18 |
Hb_170674_010 |
0.180113528 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 19 |
Hb_096819_010 |
0.1814579843 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 20 |
Hb_000046_080 |
0.1818408605 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |