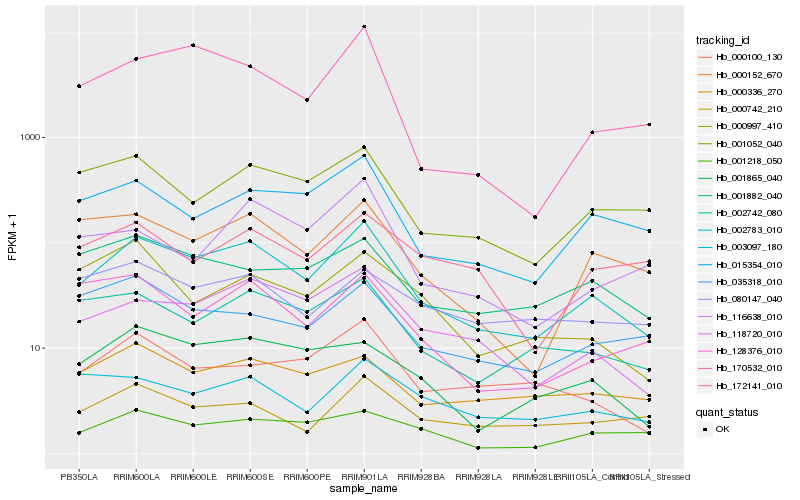
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 2 |
Hb_118720_010 |
0.1280086553 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 3 |
Hb_002742_080 |
0.14208444 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_015354_010 |
0.1492588289 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 5 |
Hb_000152_670 |
0.1524751978 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 6 |
Hb_001052_040 |
0.1537148318 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_001865_040 |
0.1557659247 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_001218_050 |
0.1647615639 |
- |
- |
Ribosome-binding protein, putative [Ricinus communis] |
| 9 |
Hb_170532_010 |
0.1655357485 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 10 |
Hb_000100_130 |
0.1659810797 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 11 |
Hb_000336_270 |
0.1673199014 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001882_040 |
0.1679295764 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 13 |
Hb_003097_180 |
0.1696152463 |
- |
- |
PREDICTED: crossover junction endonuclease MUS81 [Jatropha curcas] |
| 14 |
Hb_000742_210 |
0.1698915085 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_128376_010 |
0.1712284974 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_035318_010 |
0.1755026994 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 17 |
Hb_172141_010 |
0.1781671128 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |
| 18 |
Hb_116638_010 |
0.1802023846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000997_410 |
0.1819726154 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 20 |
Hb_080147_040 |
0.1825546607 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |