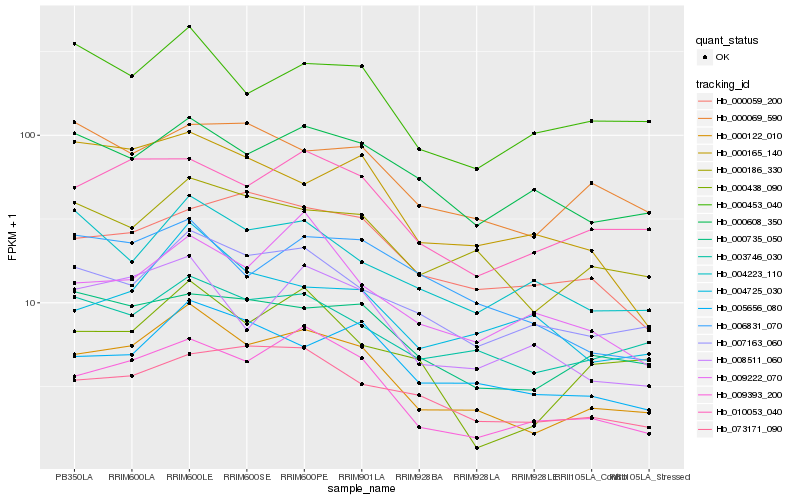
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_010 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_009393_200 |
0.1045901043 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 3 |
Hb_000186_330 |
0.1194476918 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 4 |
Hb_007163_060 |
0.1199499357 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 5 |
Hb_008511_060 |
0.1277065327 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_010053_040 |
0.1290493561 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 7 |
Hb_005656_080 |
0.1293505246 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 8 |
Hb_003746_030 |
0.1309351173 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 9 |
Hb_000735_050 |
0.1339914411 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 10 |
Hb_000453_040 |
0.138819934 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 11 |
Hb_073171_090 |
0.1388538279 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 12 |
Hb_004725_030 |
0.1448016546 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000438_090 |
0.1460576746 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 14 |
Hb_006831_070 |
0.1483195959 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000608_350 |
0.1492203478 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 16 |
Hb_000059_200 |
0.1494791987 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 17 |
Hb_004223_110 |
0.1502302082 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 18 |
Hb_000069_590 |
0.1513244471 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 19 |
Hb_000165_140 |
0.15139097 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 20 |
Hb_009222_070 |
0.1524147779 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |