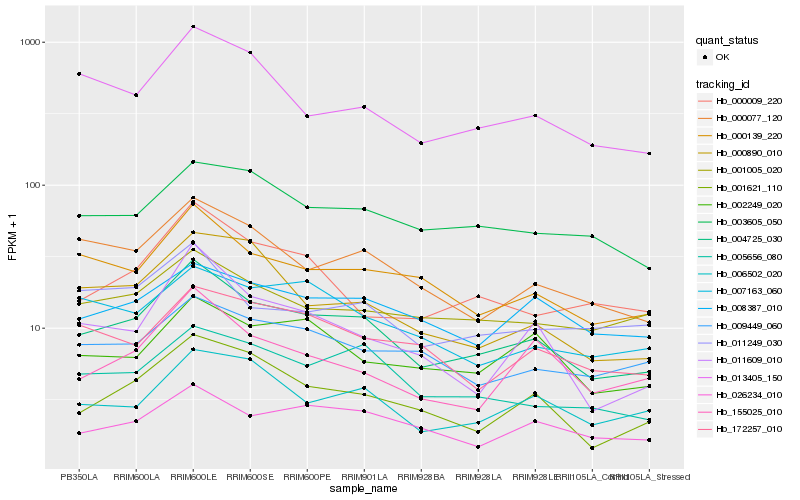
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004725_030 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000139_220 |
0.0996520373 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_005656_080 |
0.10557396 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 4 |
Hb_000077_120 |
0.1063532485 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 5 |
Hb_001621_110 |
0.1095445676 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 6 |
Hb_013405_150 |
0.1139916126 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 7 |
Hb_026234_010 |
0.1146394863 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_003605_050 |
0.1149802924 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_006502_020 |
0.1152918932 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 10 |
Hb_002249_020 |
0.118501062 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 11 |
Hb_009449_060 |
0.1193828478 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_008387_010 |
0.1211725562 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 13 |
Hb_011249_030 |
0.1220965794 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 14 |
Hb_155025_010 |
0.1222997417 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 15 |
Hb_001005_020 |
0.1225318646 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 16 |
Hb_000890_010 |
0.1236885775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_172257_010 |
0.1237874947 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 18 |
Hb_011609_010 |
0.1252958194 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 19 |
Hb_000009_220 |
0.1256515192 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_007163_060 |
0.1269693757 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |