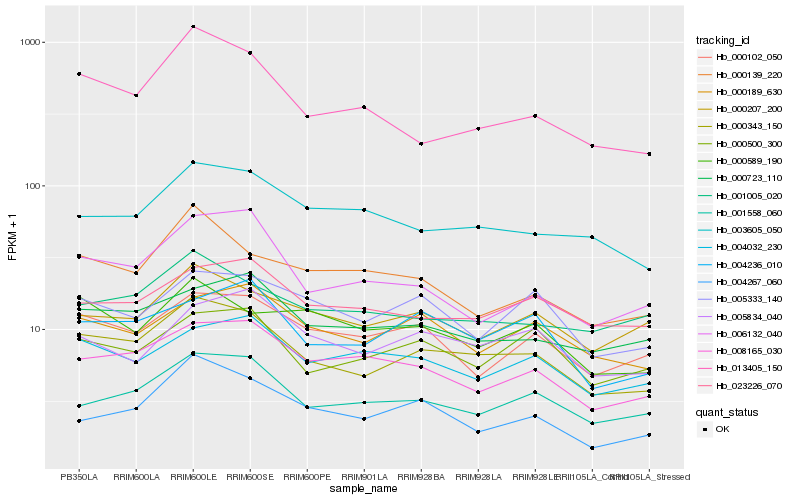
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_150 |
0.0 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 2 |
Hb_001558_060 |
0.0993581023 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 3 |
Hb_008165_030 |
0.1079497258 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 4 |
Hb_013405_150 |
0.1091946258 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 5 |
Hb_004236_010 |
0.1096061637 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 6 |
Hb_000723_110 |
0.1098824535 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_003605_050 |
0.1107777392 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000189_630 |
0.1117890774 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 9 |
Hb_000500_300 |
0.1132776274 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 10 |
Hb_000102_050 |
0.113665196 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000207_200 |
0.1137763598 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001005_020 |
0.1140558925 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 13 |
Hb_023226_070 |
0.1157234778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004032_230 |
0.1174308667 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 15 |
Hb_004267_060 |
0.1183348317 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005333_140 |
0.1184120568 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006132_040 |
0.1194407796 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 18 |
Hb_005834_040 |
0.1196721894 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 19 |
Hb_000589_190 |
0.1209553016 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000139_220 |
0.1216409015 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |