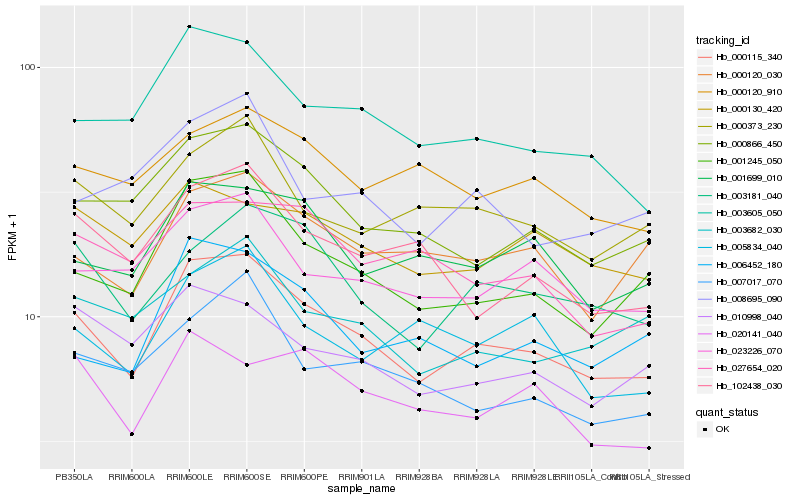
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 2 |
Hb_001245_050 |
0.0748939762 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 3 |
Hb_003605_050 |
0.0859560954 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000866_450 |
0.0895247665 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_003181_040 |
0.0936808227 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 6 |
Hb_023226_070 |
0.0937770035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001699_010 |
0.0967064707 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 8 |
Hb_000120_030 |
0.0992152301 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000373_230 |
0.099769663 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 10 |
Hb_007017_070 |
0.1026165619 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006452_180 |
0.1030230541 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 12 |
Hb_102438_030 |
0.1031764053 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003682_030 |
0.1047901876 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 14 |
Hb_000130_420 |
0.1051996348 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_010998_040 |
0.1057039504 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 16 |
Hb_005834_040 |
0.1074023862 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 17 |
Hb_020141_040 |
0.1074508069 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008695_090 |
0.107688253 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 19 |
Hb_000120_910 |
0.1085141482 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 20 |
Hb_027654_020 |
0.1098539319 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |