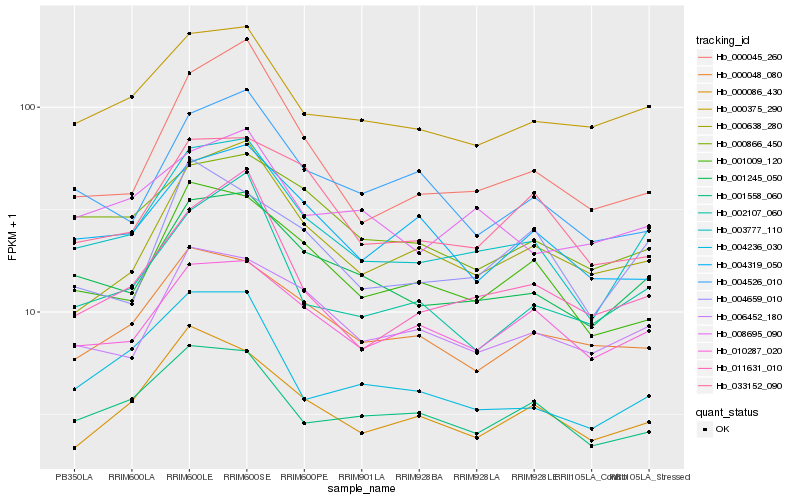
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003777_110 |
0.0 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 8 [Jatropha curcas] |
| 2 |
Hb_001245_050 |
0.0669679979 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 3 |
Hb_000375_290 |
0.0953236489 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001009_120 |
0.0989106998 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011631_010 |
0.1018257721 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 6 |
Hb_006452_180 |
0.1027724285 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 7 |
Hb_010287_020 |
0.1032528362 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 8 |
Hb_000638_280 |
0.1036949388 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 9 |
Hb_001558_060 |
0.1037325162 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000086_430 |
0.1041533057 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 11 |
Hb_000045_260 |
0.1045289642 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_002107_060 |
0.1054132032 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 13 |
Hb_004236_030 |
0.1067316826 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_000866_450 |
0.1084374158 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000048_080 |
0.1098399549 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_033152_090 |
0.1104606228 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 17 |
Hb_008695_090 |
0.111576711 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 18 |
Hb_004319_050 |
0.1122409508 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 19 |
Hb_004659_010 |
0.1130864645 |
- |
- |
Guanine nucleotide-binding subunit beta-2 [Gossypium arboreum] |
| 20 |
Hb_004526_010 |
0.1133741957 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |