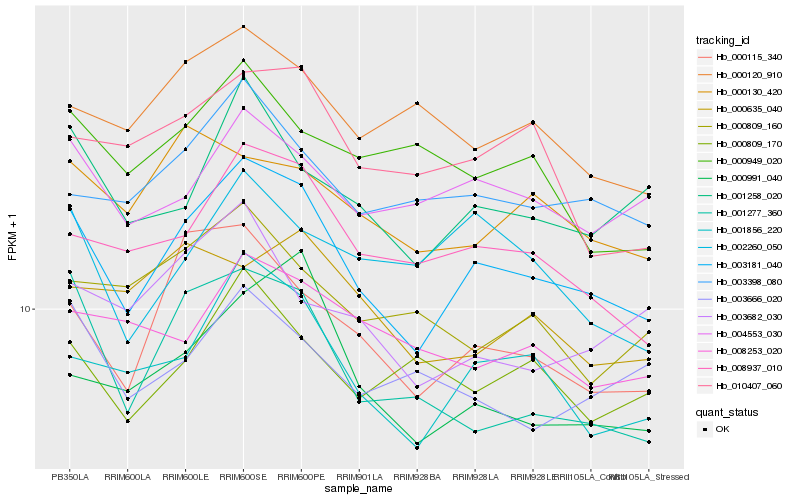
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003181_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001856_220 |
0.0808513647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008937_010 |
0.0932778214 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000115_340 |
0.0936808227 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 5 |
Hb_001258_020 |
0.1009909202 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 6 |
Hb_003398_080 |
0.1031868035 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_004553_030 |
0.1051122019 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 8 |
Hb_000991_040 |
0.1073993215 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 9 |
Hb_001277_360 |
0.1079588336 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 10 |
Hb_010407_060 |
0.1121364663 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 11 |
Hb_000130_420 |
0.1121926779 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_000120_910 |
0.1136934854 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 13 |
Hb_000809_170 |
0.1138181035 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_002260_050 |
0.1164728703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_008253_020 |
0.1171671448 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 16 |
Hb_003682_030 |
0.118298382 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 17 |
Hb_003666_020 |
0.1201309772 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000635_040 |
0.1208259336 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 19 |
Hb_000809_160 |
0.1208792682 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 20 |
Hb_000949_020 |
0.1210207267 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |