| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
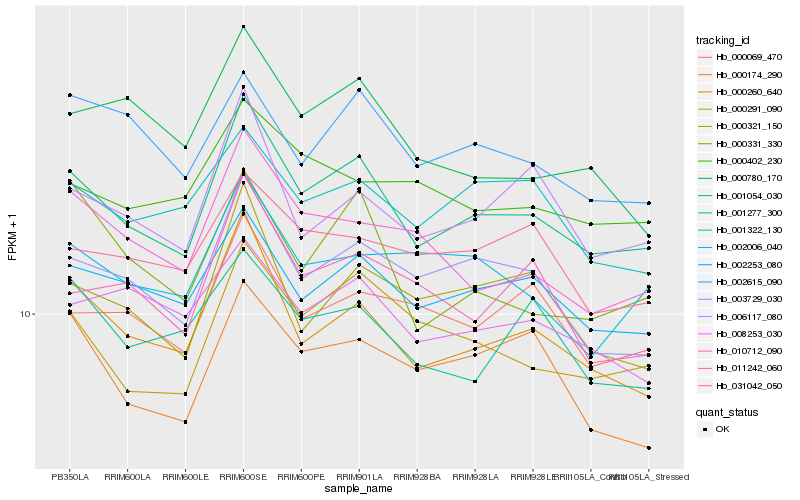
Hb_001054_030 |
0.0 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 2 |
Hb_000260_640 |
0.0570664517 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 3 |
Hb_002253_080 |
0.0660114593 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 4 |
Hb_003729_030 |
0.0676749891 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 5 |
Hb_000291_090 |
0.0727182645 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006117_080 |
0.0777637765 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000321_150 |
0.0796346493 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010712_090 |
0.0804755045 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 9 |
Hb_000069_470 |
0.0805919478 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002615_090 |
0.0831682438 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011242_060 |
0.0833853417 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 12 |
Hb_002006_040 |
0.0844343551 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 13 |
Hb_008253_030 |
0.08499798 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 14 |
Hb_001322_130 |
0.0853899558 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 15 |
Hb_031042_050 |
0.0877197779 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 16 |
Hb_000402_230 |
0.0880762532 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000331_330 |
0.0886786083 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000174_290 |
0.088896762 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_001277_300 |
0.0895516693 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 20 |
Hb_000780_170 |
0.0896256993 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |