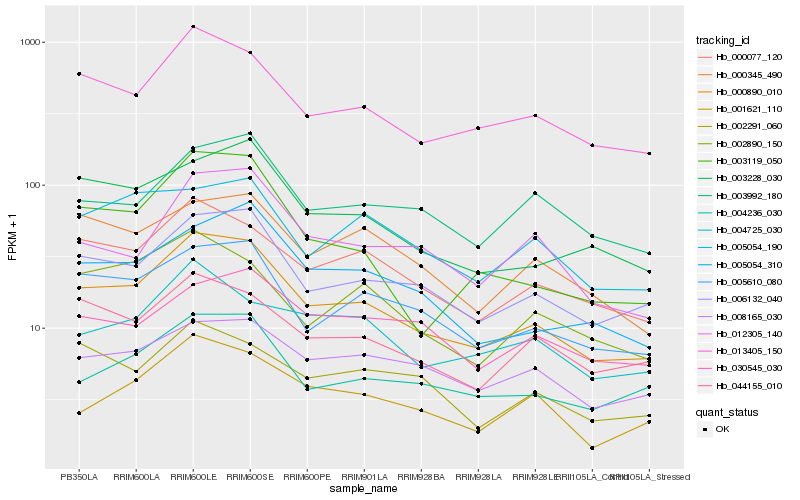
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006132_040 |
0.0765157979 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 3 |
Hb_013405_150 |
0.0802205235 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 4 |
Hb_005610_080 |
0.0892451662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000077_120 |
0.0898581389 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 6 |
Hb_004236_030 |
0.1008969675 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_044155_010 |
0.1058359824 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003228_030 |
0.1060842849 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_005054_310 |
0.10610912 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 10 |
Hb_001621_110 |
0.1122997337 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 11 |
Hb_012305_140 |
0.1123668597 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_008165_030 |
0.1152432039 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 13 |
Hb_003119_050 |
0.1153405606 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003992_180 |
0.1153859062 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000345_490 |
0.115396077 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 16 |
Hb_002890_150 |
0.1161025432 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002291_060 |
0.1201773691 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_004725_030 |
0.1236885775 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 19 |
Hb_005054_190 |
0.1255633974 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_030545_030 |
0.1272639175 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |