| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
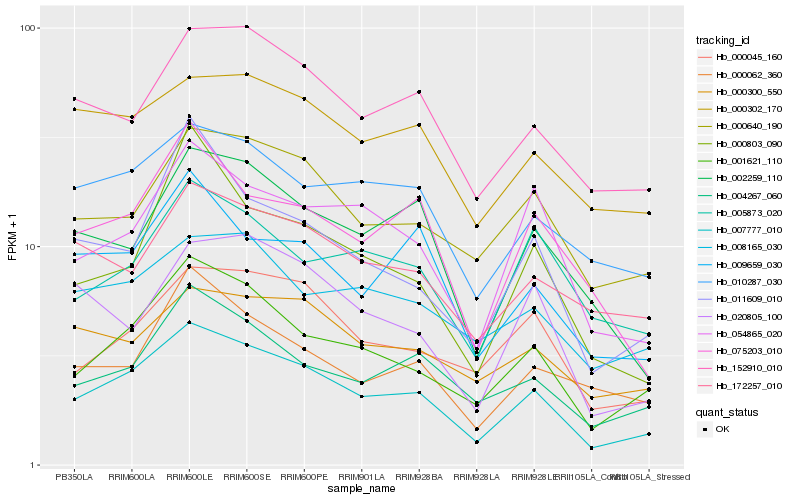
Hb_007777_010 |
0.0 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 2 |
Hb_001621_110 |
0.0805578536 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 3 |
Hb_004267_060 |
0.1114146902 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_152910_010 |
0.1141417228 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 5 |
Hb_000045_160 |
0.1150457519 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 6 |
Hb_000640_190 |
0.1153144058 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_010287_030 |
0.1167767237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_054865_020 |
0.117059237 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 9 |
Hb_009659_030 |
0.1226946217 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 10 |
Hb_002259_110 |
0.1236982567 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 11 |
Hb_011609_010 |
0.1239250075 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 12 |
Hb_075203_010 |
0.1268377296 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_172257_010 |
0.1291888584 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 14 |
Hb_008165_030 |
0.1295872979 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 15 |
Hb_000062_360 |
0.1298003295 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 16 |
Hb_000302_170 |
0.1299218316 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 17 |
Hb_000300_550 |
0.1311688418 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 18 |
Hb_020805_100 |
0.1319096012 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 19 |
Hb_000803_090 |
0.1323242623 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 20 |
Hb_005873_020 |
0.1328977092 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |