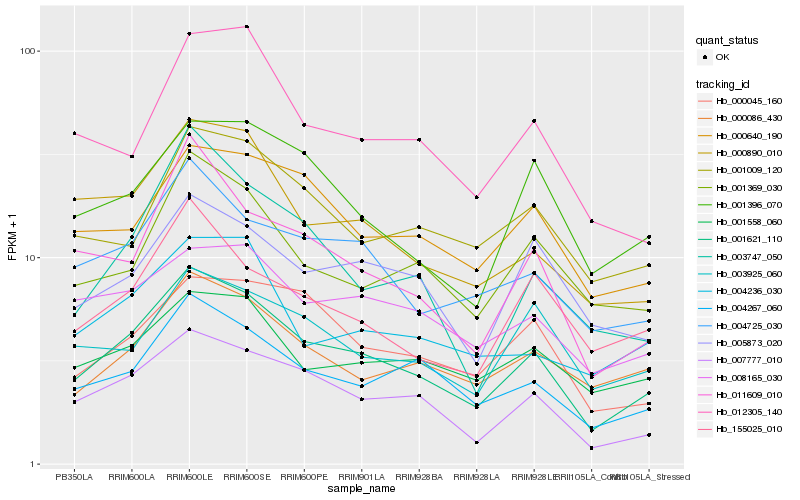
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 2 |
Hb_007777_010 |
0.0805578536 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 3 |
Hb_004267_060 |
0.1026125207 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_004725_030 |
0.1095445676 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 5 |
Hb_000890_010 |
0.1122997337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000086_430 |
0.1166940455 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 7 |
Hb_011609_010 |
0.1186218194 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 8 |
Hb_001369_030 |
0.1198978851 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000045_160 |
0.1230543308 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 10 |
Hb_000640_190 |
0.1233043457 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_155025_010 |
0.1236400058 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 12 |
Hb_001396_070 |
0.1262597315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005873_020 |
0.1262893738 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 14 |
Hb_004236_030 |
0.127152891 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_008165_030 |
0.1273514605 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 16 |
Hb_001558_060 |
0.1273672353 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001009_120 |
0.1288925436 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012305_140 |
0.1289188054 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003925_060 |
0.1296732877 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003747_050 |
0.1300113607 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |