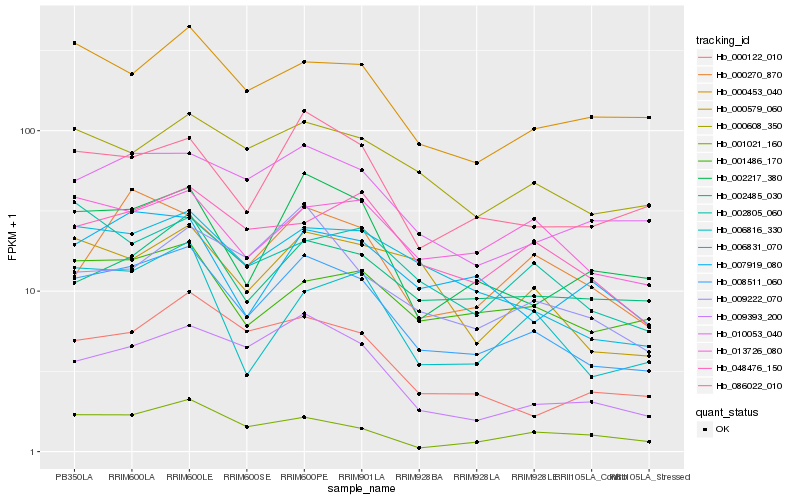
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008511_060 |
0.0 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 2 |
Hb_006831_070 |
0.1097187451 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002217_380 |
0.1190361977 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 4 |
Hb_009393_200 |
0.1195636579 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 5 |
Hb_086022_010 |
0.1199258269 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 6 |
Hb_010053_040 |
0.1253223911 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 7 |
Hb_000608_350 |
0.1275877027 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 8 |
Hb_000122_010 |
0.1277065327 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000453_040 |
0.1302357605 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 10 |
Hb_000579_060 |
0.1316744568 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001021_160 |
0.1323314835 |
- |
- |
PREDICTED: homeobox protein LUMINIDEPENDENS [Jatropha curcas] |
| 12 |
Hb_048476_150 |
0.1362597406 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_006816_330 |
0.137924494 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 14 |
Hb_001486_170 |
0.1384321863 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 15 |
Hb_009222_070 |
0.1406465024 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_002485_030 |
0.1428812606 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 17 |
Hb_007919_080 |
0.1435639785 |
- |
- |
PREDICTED: uncharacterized protein At1g76660 isoform X1 [Jatropha curcas] |
| 18 |
Hb_013726_080 |
0.1449042593 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 19 |
Hb_002805_060 |
0.1449948121 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 20 |
Hb_000270_870 |
0.1458439627 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |