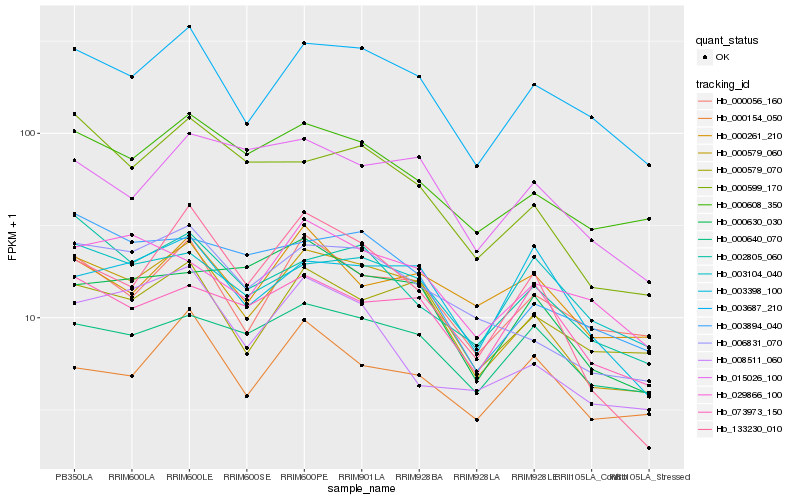
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003687_210 |
0.0892508313 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 3 |
Hb_006831_070 |
0.0936661371 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000608_350 |
0.1057376328 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 5 |
Hb_000056_160 |
0.1086451689 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000599_170 |
0.1087366688 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_000579_070 |
0.1102058297 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 8 |
Hb_002805_060 |
0.1146161158 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 9 |
Hb_003894_040 |
0.1153540181 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 10 |
Hb_133230_010 |
0.1203725726 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 11 |
Hb_000640_070 |
0.128726691 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 12 |
Hb_015026_100 |
0.1304576907 |
- |
- |
PREDICTED: transmembrane protein 184C-like [Jatropha curcas] |
| 13 |
Hb_073973_150 |
0.1311046113 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000630_030 |
0.1311503281 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 15 |
Hb_008511_060 |
0.1316744568 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 16 |
Hb_003104_040 |
0.1338038482 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 17 |
Hb_000154_050 |
0.135398158 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 18 |
Hb_029866_100 |
0.1368734545 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 19 |
Hb_000261_210 |
0.138173746 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_003398_100 |
0.1388314583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |