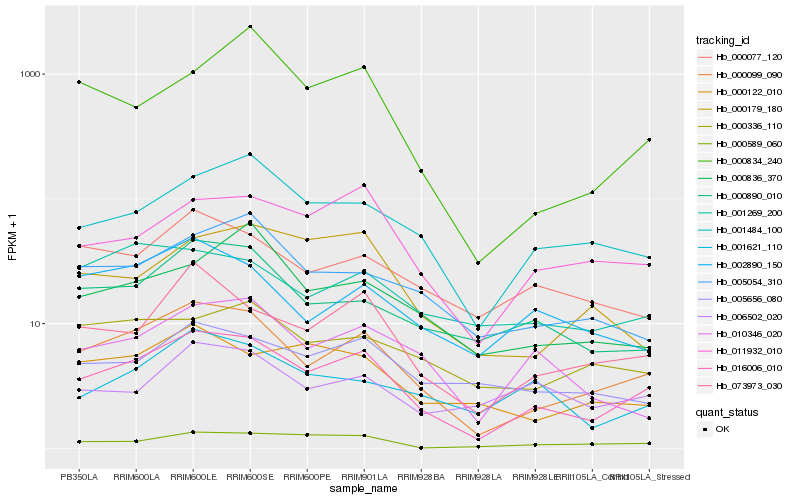
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016006_010 |
0.0 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000099_090 |
0.0802999652 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 3 |
Hb_073973_030 |
0.1395853223 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 4 |
Hb_011932_010 |
0.1432946693 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 5 |
Hb_001484_100 |
0.1481907912 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 6 |
Hb_000589_060 |
0.1633299594 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 7 |
Hb_000890_010 |
0.1646852718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001621_110 |
0.1696521141 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 9 |
Hb_010346_020 |
0.1696712317 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 10 |
Hb_000077_120 |
0.171633367 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 11 |
Hb_005656_080 |
0.1722980247 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 12 |
Hb_005054_310 |
0.1748950917 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 13 |
Hb_002890_150 |
0.1764465717 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000122_010 |
0.1772540705 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000836_370 |
0.1823460213 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001269_200 |
0.1826659835 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 17 |
Hb_000834_240 |
0.1833640392 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 18 |
Hb_000179_180 |
0.1836222007 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 19 |
Hb_006502_020 |
0.1837516248 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 20 |
Hb_000336_110 |
0.1838702653 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |