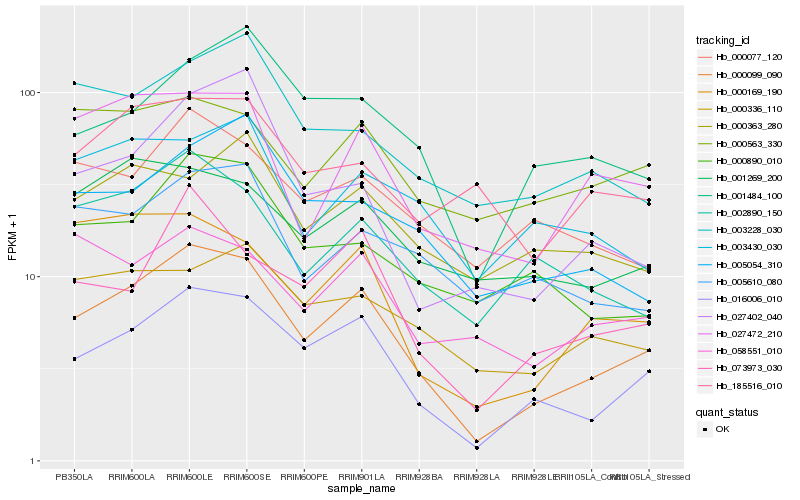
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_090 |
0.0 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 2 |
Hb_016006_010 |
0.0802999652 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_073973_030 |
0.1453878247 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 4 |
Hb_027472_210 |
0.1520523207 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 5 |
Hb_002890_150 |
0.1569656323 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000169_190 |
0.1575896958 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 7 |
Hb_000890_010 |
0.1623548831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001484_100 |
0.1628700477 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 9 |
Hb_027402_040 |
0.1664795656 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 10 |
Hb_005054_310 |
0.1672867073 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 11 |
Hb_000077_120 |
0.1673990062 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 12 |
Hb_003228_030 |
0.1683865899 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_000336_110 |
0.1692775531 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_005610_080 |
0.1699168876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001269_200 |
0.1715508215 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 16 |
Hb_058551_010 |
0.1741610669 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_003430_030 |
0.1770555564 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 18 |
Hb_000563_330 |
0.1784453502 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000363_280 |
0.1788179641 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 20 |
Hb_185516_010 |
0.1797278751 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |