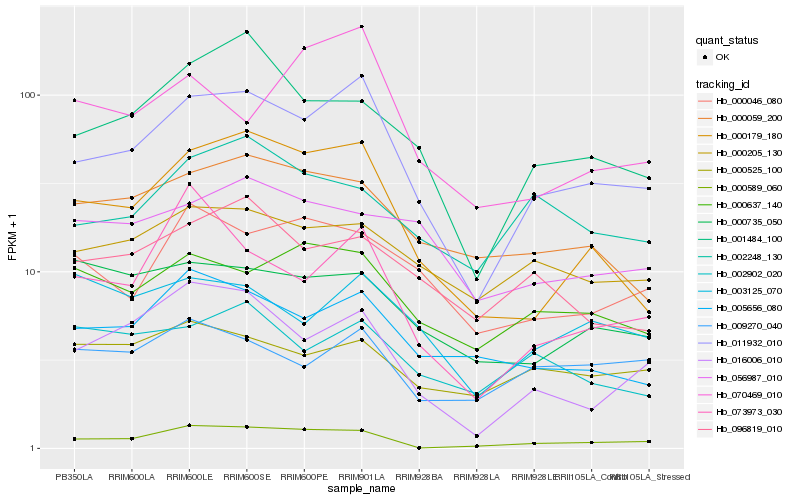
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011932_010 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 2 |
Hb_000179_180 |
0.1268311752 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 3 |
Hb_016006_010 |
0.1432946693 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000589_060 |
0.1463453092 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 5 |
Hb_001484_100 |
0.1481228153 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 6 |
Hb_005656_080 |
0.1493489749 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 7 |
Hb_000205_130 |
0.1549675003 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000637_140 |
0.1570784715 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 9 |
Hb_000059_200 |
0.157503957 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 10 |
Hb_096819_010 |
0.1624333031 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000046_080 |
0.1625319434 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 12 |
Hb_073973_030 |
0.164811293 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002248_130 |
0.1688822586 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 14 |
Hb_009270_040 |
0.1691271333 |
- |
- |
- |
| 15 |
Hb_003125_070 |
0.1702675817 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 16 |
Hb_002902_020 |
0.1704143569 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 17 |
Hb_000525_100 |
0.1729431424 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 18 |
Hb_070469_010 |
0.1734052874 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 19 |
Hb_056987_010 |
0.174978317 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 20 |
Hb_000735_050 |
0.175169571 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |