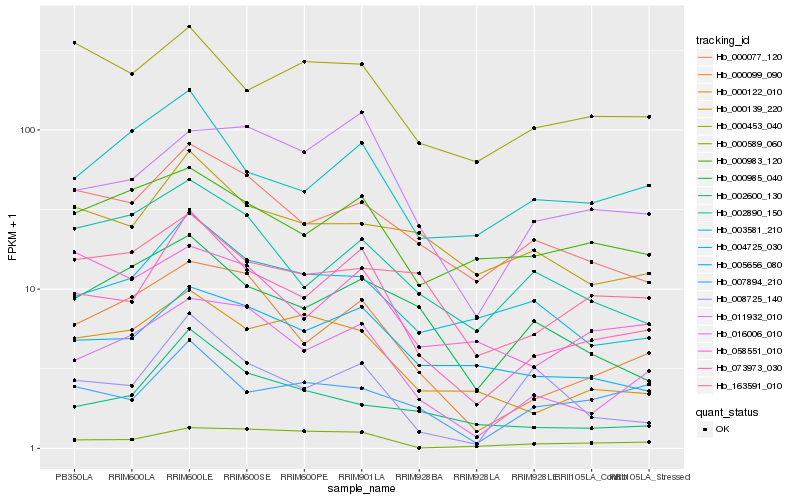
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073973_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 2 |
Hb_016006_010 |
0.1395853223 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_000099_090 |
0.1453878247 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 4 |
Hb_005656_080 |
0.1628082417 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 5 |
Hb_000077_120 |
0.16362209 |
- |
- |
PREDICTED: two pore calcium channel protein 1 [Jatropha curcas] |
| 6 |
Hb_003581_210 |
0.1644682515 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011932_010 |
0.164811293 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 8 |
Hb_000122_010 |
0.1700149764 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_004725_030 |
0.1731470901 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 10 |
Hb_008725_140 |
0.1734008163 |
- |
- |
PREDICTED: kynurenine--oxoglutarate transaminase-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002600_130 |
0.1795684105 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 12 |
Hb_007894_210 |
0.1807068637 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 13 |
Hb_000985_040 |
0.1827675641 |
transcription factor |
TF Family: Orphans |
B-box type zinc finger protein with CCT domain isoform 1 [Theobroma cacao] |
| 14 |
Hb_002890_150 |
0.1828023222 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000139_220 |
0.18312174 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000589_060 |
0.1839823362 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 17 |
Hb_163591_010 |
0.1873799568 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000453_040 |
0.1883412287 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 19 |
Hb_058551_010 |
0.1885408692 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000983_120 |
0.190082847 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |