| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
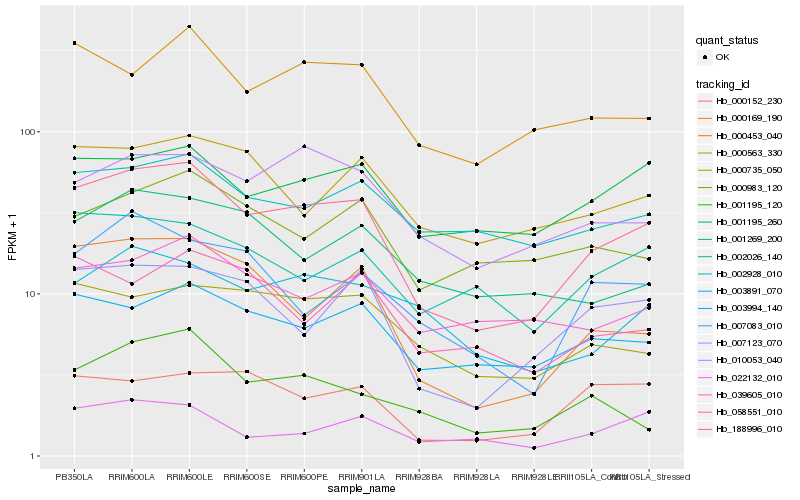
Hb_188996_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000169_190 |
0.1258174677 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 3 |
Hb_007123_070 |
0.1296084868 |
- |
- |
- |
| 4 |
Hb_000453_040 |
0.1341383965 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 5 |
Hb_003994_140 |
0.1346827599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010053_040 |
0.1396127426 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 7 |
Hb_003891_070 |
0.1405125999 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 8 |
Hb_001195_260 |
0.1420828803 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 9 |
Hb_000735_050 |
0.144091663 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 10 |
Hb_002026_140 |
0.1452951668 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 11 |
Hb_000563_330 |
0.1459681083 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_007083_010 |
0.1472428157 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 13-like, partial [Nelumbo nucifera] |
| 13 |
Hb_002928_010 |
0.1476042514 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 14 |
Hb_039605_010 |
0.1478104407 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 15 |
Hb_022132_010 |
0.1485176497 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_000152_230 |
0.1509259043 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 17 |
Hb_001269_200 |
0.1513119408 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000983_120 |
0.1522285964 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_058551_010 |
0.1523667581 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_001195_120 |
0.1524422944 |
- |
- |
oxidoreductase, putative [Ricinus communis] |