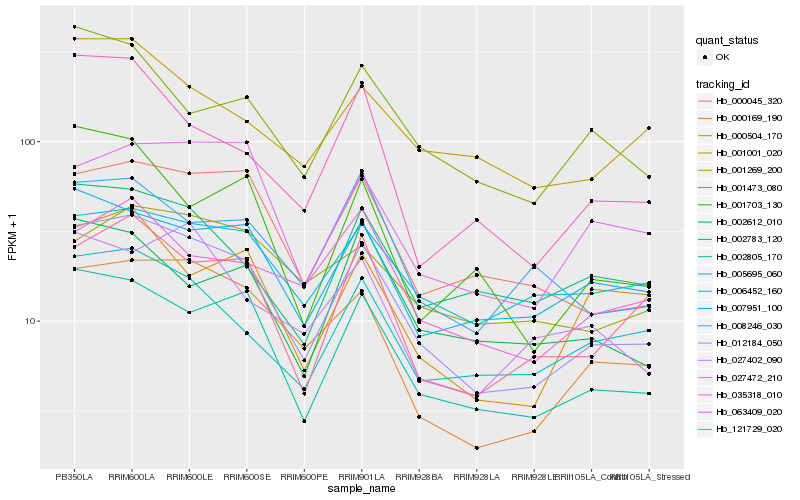
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012184_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 2 |
Hb_002805_170 |
0.0864541848 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 3 |
Hb_000169_190 |
0.1011779448 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 4 |
Hb_000045_320 |
0.1095744053 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 5 |
Hb_121729_020 |
0.1163708178 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_007951_100 |
0.1224459042 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_005695_060 |
0.1242664215 |
- |
- |
PREDICTED: probable tRNA modification GTPase MnmE [Jatropha curcas] |
| 8 |
Hb_027472_210 |
0.1271513391 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 9 |
Hb_002783_120 |
0.1342437291 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 10 |
Hb_008246_030 |
0.1344327854 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 11 |
Hb_006452_160 |
0.1347476554 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 12 |
Hb_063409_020 |
0.1350848383 |
- |
- |
Calcium-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000504_170 |
0.1360571043 |
- |
- |
hypothetical protein B456_006G033900 [Gossypium raimondii] |
| 14 |
Hb_035318_010 |
0.138797541 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 15 |
Hb_001703_130 |
0.1394664822 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like [Populus euphratica] |
| 16 |
Hb_001001_020 |
0.1404454298 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Populus euphratica] |
| 17 |
Hb_001473_080 |
0.1412005295 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 18 |
Hb_027402_090 |
0.1414031438 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 19 |
Hb_002612_010 |
0.1421816815 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 20 |
Hb_001269_200 |
0.1454740272 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |