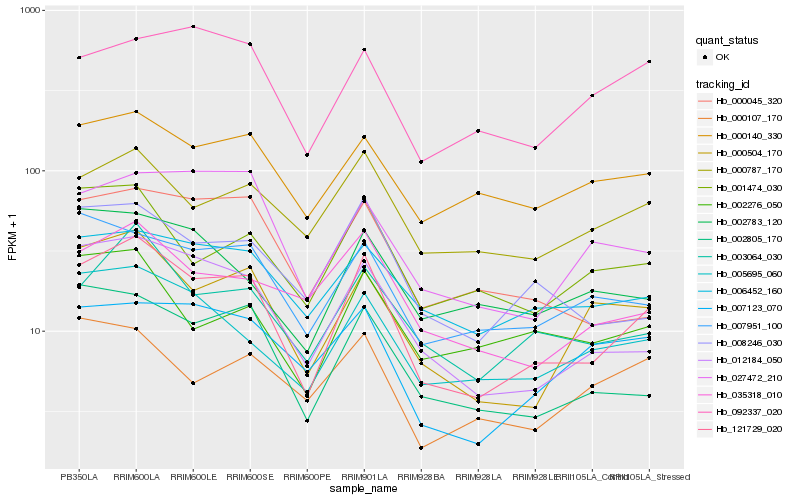
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121729_020 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_012184_050 |
0.1163708178 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 3 |
Hb_000504_170 |
0.1267804911 |
- |
- |
hypothetical protein B456_006G033900 [Gossypium raimondii] |
| 4 |
Hb_003064_030 |
0.1277066627 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_002805_170 |
0.1277799626 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 6 |
Hb_005695_060 |
0.1331567779 |
- |
- |
PREDICTED: probable tRNA modification GTPase MnmE [Jatropha curcas] |
| 7 |
Hb_008246_030 |
0.1387919013 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 8 |
Hb_007951_100 |
0.1409729821 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_035318_010 |
0.1424641758 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 10 |
Hb_001474_030 |
0.1427187196 |
- |
- |
PREDICTED: uncharacterized protein LOC105650916 [Jatropha curcas] |
| 11 |
Hb_006452_160 |
0.1435285741 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 12 |
Hb_000787_170 |
0.1444175984 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 13 |
Hb_000045_320 |
0.1470714675 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000140_330 |
0.1487106888 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 15 |
Hb_027472_210 |
0.1489626396 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 16 |
Hb_007123_070 |
0.1491223805 |
- |
- |
- |
| 17 |
Hb_002276_050 |
0.1544968858 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 18 |
Hb_092337_020 |
0.1579637654 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 19 |
Hb_002783_120 |
0.1595168882 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 20 |
Hb_000107_170 |
0.1612105271 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |