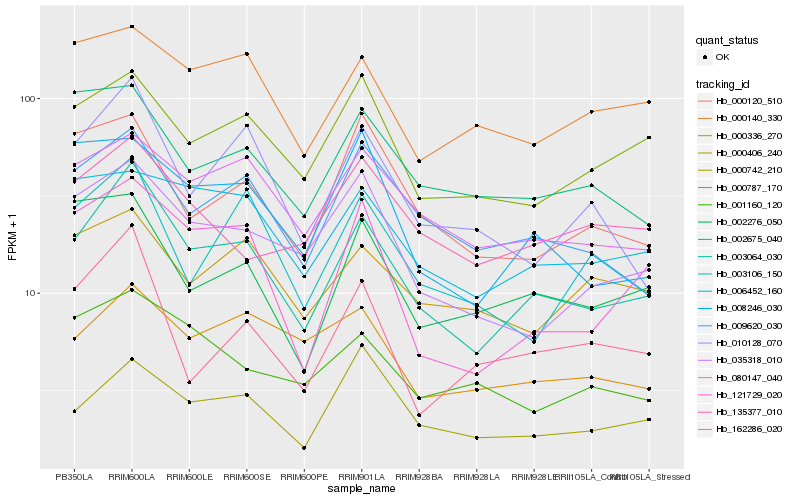
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003064_030 |
0.0 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_009620_030 |
0.1227479774 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 3 |
Hb_080147_040 |
0.1249730949 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 4 |
Hb_035318_010 |
0.1268282163 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 5 |
Hb_121729_020 |
0.1277066627 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000787_170 |
0.1299646227 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 7 |
Hb_010128_070 |
0.1326779233 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_006452_160 |
0.1356158892 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 9 |
Hb_000742_210 |
0.1386420151 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 10 |
Hb_008246_030 |
0.1416094379 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 11 |
Hb_000336_270 |
0.1419274799 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000120_510 |
0.1419969328 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002276_050 |
0.1425891252 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000140_330 |
0.1428428551 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 15 |
Hb_162286_020 |
0.1443473262 |
- |
- |
PREDICTED: uncharacterized protein LOC105642702 [Jatropha curcas] |
| 16 |
Hb_002675_040 |
0.1443755103 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 17 |
Hb_003106_150 |
0.1448015838 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 18 |
Hb_001160_120 |
0.1455104991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000406_240 |
0.1474490481 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_135377_010 |
0.1478316603 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |