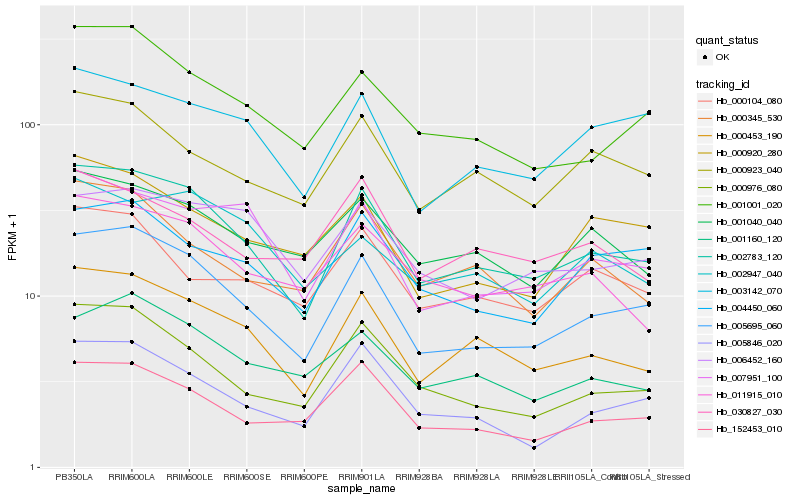
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 2 |
Hb_005695_060 |
0.0537606888 |
- |
- |
PREDICTED: probable tRNA modification GTPase MnmE [Jatropha curcas] |
| 3 |
Hb_000453_190 |
0.0615670258 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 4 |
Hb_000976_080 |
0.094636247 |
- |
- |
hypothetical protein VITISV_034376 [Vitis vinifera] |
| 5 |
Hb_001001_020 |
0.0982312185 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Populus euphratica] |
| 6 |
Hb_011915_010 |
0.1025484056 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 7 |
Hb_001160_120 |
0.1039658413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_152453_010 |
0.104393578 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001040_040 |
0.1072813496 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 10 |
Hb_007951_100 |
0.107980924 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_005846_020 |
0.1094414393 |
- |
- |
PREDICTED: tubulin-folding cofactor C [Jatropha curcas] |
| 12 |
Hb_000923_040 |
0.1095310444 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 13 |
Hb_000345_530 |
0.1095367241 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003142_070 |
0.115730702 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 15 |
Hb_006452_160 |
0.1191716147 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 16 |
Hb_004450_060 |
0.12016063 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002947_040 |
0.1204465286 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |
| 18 |
Hb_030827_030 |
0.1218404163 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 19 |
Hb_000104_080 |
0.1234705747 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 20 |
Hb_000920_280 |
0.1235237459 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |