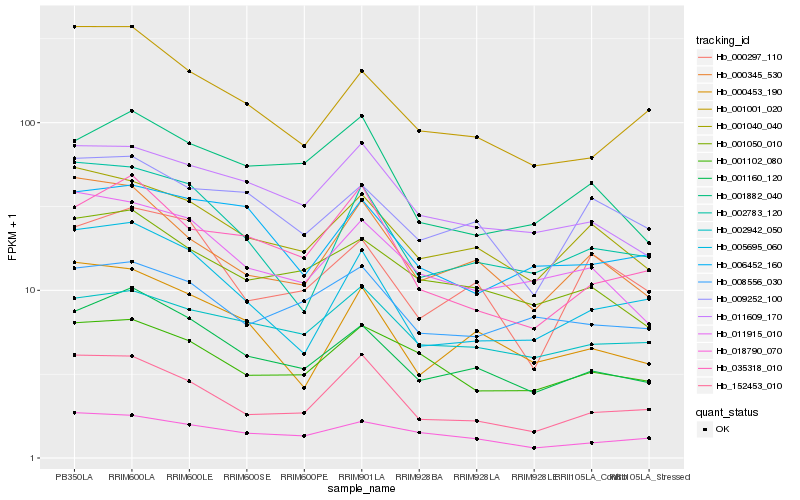
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001050_010 |
0.0833714278 |
- |
- |
Advillin [Gossypium arboreum] |
| 3 |
Hb_001040_040 |
0.0938584612 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 4 |
Hb_011915_010 |
0.0964614065 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 5 |
Hb_001001_020 |
0.0978246298 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Populus euphratica] |
| 6 |
Hb_011609_170 |
0.1014035309 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 7 |
Hb_002783_120 |
0.1039658413 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 8 |
Hb_005695_060 |
0.104646931 |
- |
- |
PREDICTED: probable tRNA modification GTPase MnmE [Jatropha curcas] |
| 9 |
Hb_000453_190 |
0.1055521566 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 10 |
Hb_000297_110 |
0.1076196044 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 11 |
Hb_018790_070 |
0.1076335803 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 12 |
Hb_009252_100 |
0.1100384514 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 13 |
Hb_000345_530 |
0.1111256719 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 14 |
Hb_152453_010 |
0.1122894564 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001102_080 |
0.1147027615 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_006452_160 |
0.1149617666 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 17 |
Hb_035318_010 |
0.115864155 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 18 |
Hb_008556_030 |
0.116397612 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001882_040 |
0.1165991004 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 20 |
Hb_002942_050 |
0.1169250644 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |