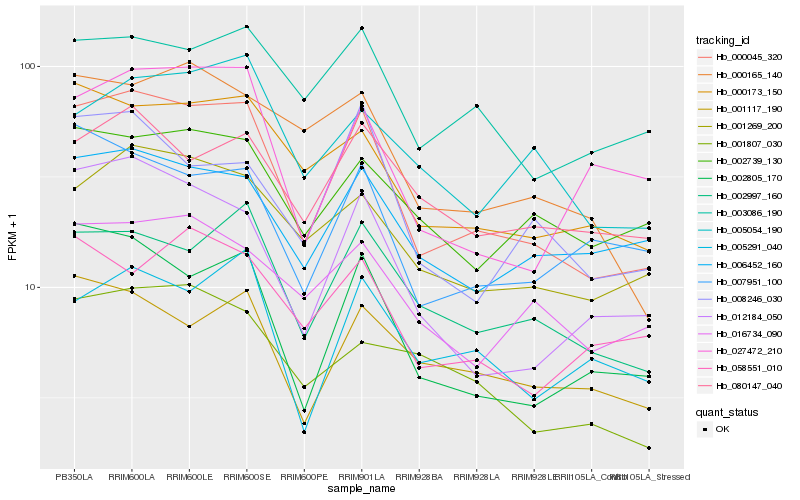
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000045_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002805_170 |
0.1029559428 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 3 |
Hb_002997_160 |
0.106796597 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000173_150 |
0.1070503647 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 5 |
Hb_012184_050 |
0.1095744053 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 6 |
Hb_001269_200 |
0.1119181371 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 7 |
Hb_001117_190 |
0.1247829555 |
transcription factor |
TF Family: FAR1 |
FAR1-related sequence 11 [Theobroma cacao] |
| 8 |
Hb_006452_160 |
0.1259281145 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 9 |
Hb_008246_030 |
0.1267614693 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 10 |
Hb_007951_100 |
0.1268575891 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_016734_090 |
0.1275958952 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 12 |
Hb_003086_190 |
0.1283794865 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 13 |
Hb_000165_140 |
0.1295864979 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 14 |
Hb_005291_040 |
0.1305084465 |
- |
- |
PREDICTED: gamma-gliadin [Jatropha curcas] |
| 15 |
Hb_027472_210 |
0.1306804788 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 16 |
Hb_002739_130 |
0.1326241399 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 17 |
Hb_058551_010 |
0.1336982913 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_080147_040 |
0.1337316505 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 19 |
Hb_001807_030 |
0.135196835 |
- |
- |
PREDICTED: CDT1-like protein b [Jatropha curcas] |
| 20 |
Hb_005054_190 |
0.1359264536 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |