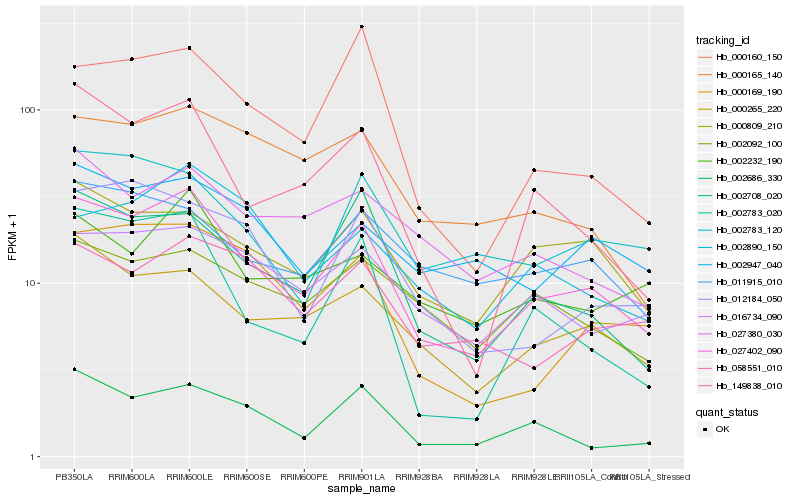
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027402_090 |
0.0 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 2 |
Hb_000265_220 |
0.1162690118 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 3 |
Hb_002708_020 |
0.1196256366 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_149838_010 |
0.1256410495 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 5 |
Hb_016734_090 |
0.1317185818 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000809_210 |
0.1329354155 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 7 |
Hb_000169_190 |
0.1362333878 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 8 |
Hb_000165_140 |
0.1385284535 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 9 |
Hb_002686_330 |
0.1387804854 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002783_020 |
0.1404168441 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X4 [Jatropha curcas] |
| 11 |
Hb_058551_010 |
0.140774638 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_011915_010 |
0.1409417896 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 13 |
Hb_000160_150 |
0.1411668018 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 14 |
Hb_027380_030 |
0.1413602842 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_012184_050 |
0.1414031438 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 16 |
Hb_002232_190 |
0.1418829507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002783_120 |
0.1422503291 |
- |
- |
PREDICTED: uncharacterized protein LOC105635329 [Jatropha curcas] |
| 18 |
Hb_002947_040 |
0.1431088405 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |
| 19 |
Hb_002890_150 |
0.1446213392 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002092_100 |
0.1458929113 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |