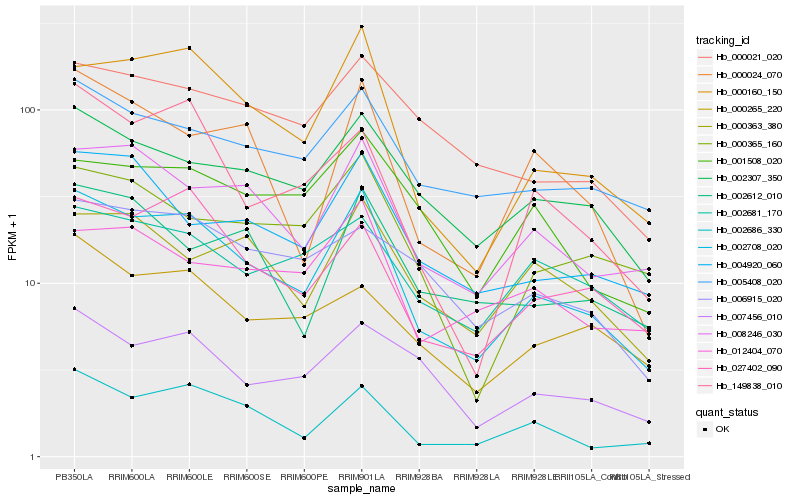
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002708_020 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000160_150 |
0.0948614184 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 3 |
Hb_002686_330 |
0.1116569602 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_008246_030 |
0.1144246396 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 5 |
Hb_027402_090 |
0.1196256366 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 6 |
Hb_002307_350 |
0.1261404647 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_005408_020 |
0.1302412999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004920_060 |
0.1333443271 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 9 |
Hb_149838_010 |
0.1337601434 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 10 |
Hb_000024_070 |
0.1370409438 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007456_010 |
0.1404204845 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 12 |
Hb_000365_160 |
0.144223722 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 13 |
Hb_000363_380 |
0.1455750059 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 14 |
Hb_000265_220 |
0.1467286013 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 15 |
Hb_002681_170 |
0.1471566996 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 16 |
Hb_000021_020 |
0.1472991893 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Malus domestica] |
| 17 |
Hb_002612_010 |
0.1478337284 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 18 |
Hb_012404_070 |
0.1481400395 |
- |
- |
PREDICTED: uncharacterized protein LOC105629465 [Jatropha curcas] |
| 19 |
Hb_006915_020 |
0.1498408345 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 20 |
Hb_001508_020 |
0.150031704 |
- |
- |
myosin vIII, putative [Ricinus communis] |