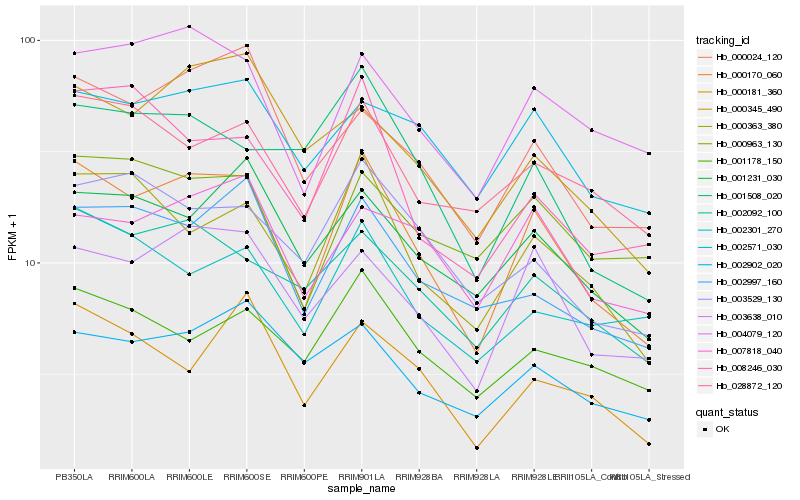
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_060 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 2 |
Hb_003638_010 |
0.1093099004 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 3 |
Hb_000363_380 |
0.1134654697 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 4 |
Hb_004079_120 |
0.1218829441 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 5 |
Hb_002092_100 |
0.1233959822 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 6 |
Hb_000024_120 |
0.1298569218 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 7 |
Hb_002997_160 |
0.1314247241 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001231_030 |
0.1317539989 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 9 |
Hb_028872_120 |
0.132968526 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 10 |
Hb_003529_130 |
0.1343846982 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_002571_030 |
0.1345698486 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 12 |
Hb_000963_130 |
0.1361493624 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001178_150 |
0.1371780831 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 14 |
Hb_007818_040 |
0.1385103447 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 15 |
Hb_008246_030 |
0.1390020314 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 16 |
Hb_001508_020 |
0.139047936 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 17 |
Hb_002902_020 |
0.1401613372 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 18 |
Hb_002301_270 |
0.1407192399 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 19 |
Hb_000181_360 |
0.1409914635 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 20 |
Hb_000345_490 |
0.141122288 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |