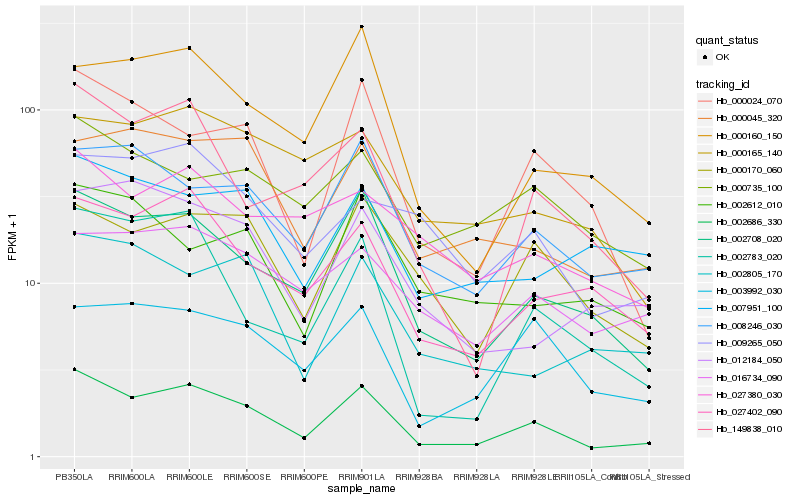
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_330 |
0.0 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002708_020 |
0.1116569602 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_027402_090 |
0.1387804854 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 4 |
Hb_000024_070 |
0.1452515628 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_008246_030 |
0.1453111517 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 6 |
Hb_000170_060 |
0.1499974404 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 7 |
Hb_149838_010 |
0.1520196492 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 8 |
Hb_000045_320 |
0.1528921661 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003992_030 |
0.1540383449 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic [Gossypium arboreum] |
| 10 |
Hb_002783_020 |
0.1547075802 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000160_150 |
0.1586684686 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 12 |
Hb_016734_090 |
0.1662521378 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 13 |
Hb_002805_170 |
0.1664916447 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 14 |
Hb_012184_050 |
0.1675196569 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 15 |
Hb_009265_050 |
0.1702615997 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 16 |
Hb_007951_100 |
0.1703373717 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 17 |
Hb_027380_030 |
0.1710844829 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000735_100 |
0.1722229215 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 19 |
Hb_000165_140 |
0.1722458849 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |
| 20 |
Hb_002612_010 |
0.1757378908 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |