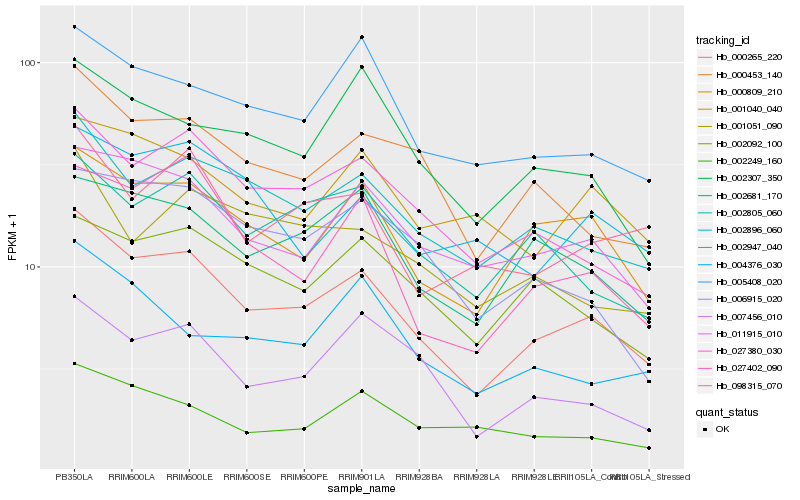
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000265_220 |
0.0 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 2 |
Hb_000809_210 |
0.0983732038 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 3 |
Hb_002896_060 |
0.0996955025 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 4 |
Hb_027380_030 |
0.1003115716 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_011915_010 |
0.1032293021 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 6 |
Hb_000453_140 |
0.1056765835 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 7 |
Hb_002681_170 |
0.1145659256 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 8 |
Hb_027402_090 |
0.1162690118 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 9 |
Hb_005408_020 |
0.1186945668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002092_100 |
0.120078224 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 11 |
Hb_007456_010 |
0.121111134 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 12 |
Hb_002805_060 |
0.1230945529 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 13 |
Hb_006915_020 |
0.1236972771 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 14 |
Hb_098315_070 |
0.1268048707 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001040_040 |
0.1273083143 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 16 |
Hb_002307_350 |
0.1274549056 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 17 |
Hb_002249_160 |
0.1292779383 |
- |
- |
- |
| 18 |
Hb_004376_030 |
0.1321472705 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 19 |
Hb_001051_090 |
0.1323891609 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 20 |
Hb_002947_040 |
0.1326198244 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |