| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
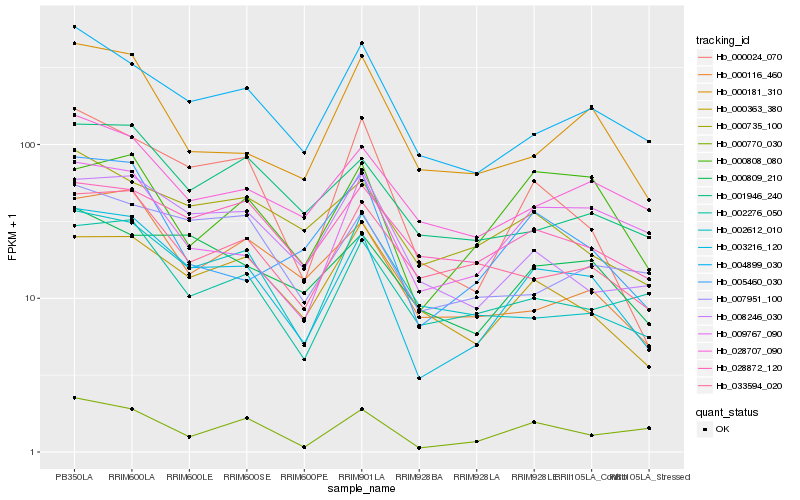
Hb_003216_120 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004899_030 |
0.1324166305 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000024_070 |
0.132765242 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000809_210 |
0.1390002591 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 5 |
Hb_028707_090 |
0.1492199896 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 6 |
Hb_000363_380 |
0.1497988217 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 7 |
Hb_008246_030 |
0.1511548651 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 8 |
Hb_002276_050 |
0.151163907 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000770_030 |
0.1517780267 |
- |
- |
- |
| 10 |
Hb_009767_090 |
0.1520328639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001946_240 |
0.1523064525 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 12 |
Hb_005460_030 |
0.1524937212 |
- |
- |
PREDICTED: RING-H2 finger protein ATL65 [Jatropha curcas] |
| 13 |
Hb_000116_460 |
0.1531693931 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 14 |
Hb_007951_100 |
0.1531909247 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000735_100 |
0.1531956195 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 16 |
Hb_000181_310 |
0.1538400975 |
- |
- |
hypothetical protein JCGZ_20998 [Jatropha curcas] |
| 17 |
Hb_033594_020 |
0.1545521977 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 18 |
Hb_000808_080 |
0.1556794575 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 3 [Jatropha curcas] |
| 19 |
Hb_002612_010 |
0.1571388324 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 20 |
Hb_028872_120 |
0.1579065987 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |