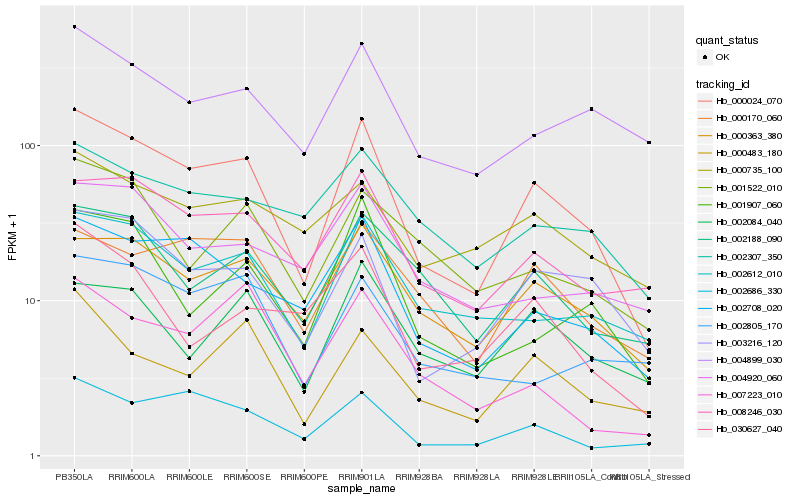
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000024_070 |
0.0 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003216_120 |
0.132765242 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000363_380 |
0.1347111291 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 4 |
Hb_002708_020 |
0.1370409438 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002612_010 |
0.1432529545 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 6 |
Hb_008246_030 |
0.1443181477 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 7 |
Hb_002686_330 |
0.1452515628 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000170_060 |
0.153794332 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 9 |
Hb_002188_090 |
0.156476715 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 10 |
Hb_000483_180 |
0.1582233427 |
- |
- |
- |
| 11 |
Hb_001907_060 |
0.1603747086 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 12 |
Hb_004899_030 |
0.160807498 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002084_040 |
0.1661908864 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002307_350 |
0.1719537755 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_001522_010 |
0.1729727554 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 16 |
Hb_030627_040 |
0.1752771047 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 17 |
Hb_004920_060 |
0.1754482875 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 18 |
Hb_002805_170 |
0.1756822612 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 19 |
Hb_000735_100 |
0.1780777761 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 20 |
Hb_007223_010 |
0.1786142389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |