| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
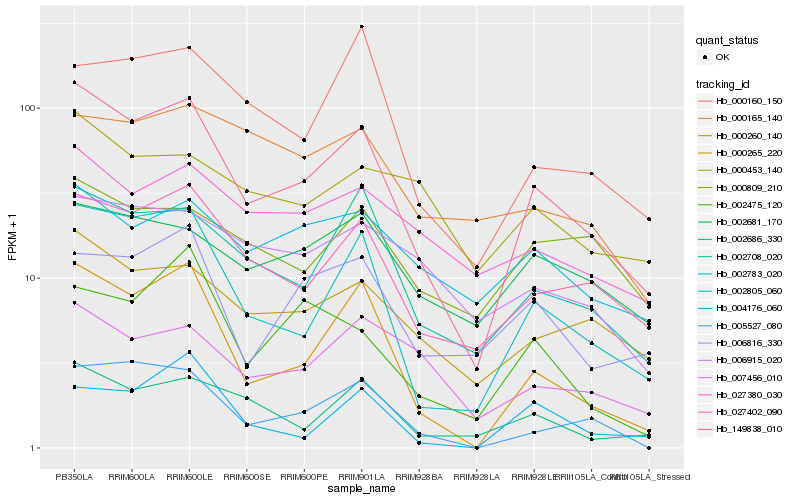
Hb_149838_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 2 |
Hb_002783_020 |
0.1050930832 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000260_140 |
0.1166292392 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 4 |
Hb_027402_090 |
0.1256410495 |
- |
- |
endonuclease, putative [Ricinus communis] |
| 5 |
Hb_002708_020 |
0.1337601434 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000265_220 |
0.1482207801 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 7 |
Hb_002686_330 |
0.1520196492 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005527_080 |
0.157793714 |
- |
- |
hypothetical protein CICLE_v10023951mg, partial [Citrus clementina] |
| 9 |
Hb_006816_330 |
0.1711265166 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 10 |
Hb_027380_030 |
0.1738218919 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_007456_010 |
0.1748391081 |
- |
- |
hypothetical protein L484_003540 [Morus notabilis] |
| 12 |
Hb_000160_150 |
0.1768396872 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 13 |
Hb_002805_060 |
0.1838893828 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 14 |
Hb_002681_170 |
0.1843665602 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 15 |
Hb_006915_020 |
0.1844451573 |
- |
- |
PREDICTED: formamidopyrimidine-DNA glycosylase isoform X1 [Jatropha curcas] |
| 16 |
Hb_002475_120 |
0.1852123275 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 17 |
Hb_000809_210 |
0.1897205396 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 18 |
Hb_000453_140 |
0.1907565682 |
- |
- |
hypothetical protein POPTR_0009s02600g [Populus trichocarpa] |
| 19 |
Hb_004176_060 |
0.195771325 |
- |
- |
- |
| 20 |
Hb_000165_140 |
0.1959592327 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 isoform X2 [Populus euphratica] |