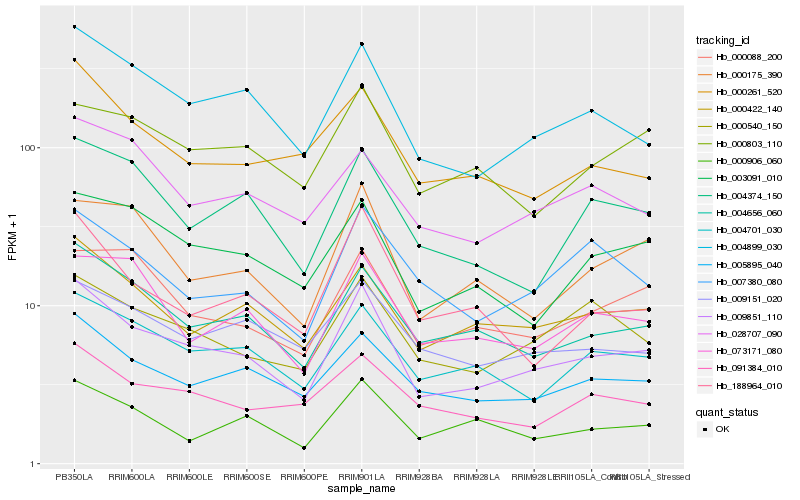
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009851_110 |
0.0 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 2 |
Hb_004899_030 |
0.0911204298 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004656_060 |
0.1008251451 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 4 |
Hb_005895_040 |
0.1043430955 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 5 |
Hb_000422_140 |
0.1079017421 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004701_030 |
0.1080921315 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 7 |
Hb_003091_010 |
0.1179002842 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 8 |
Hb_000540_150 |
0.1199444501 |
- |
- |
PREDICTED: uncharacterized protein YKR070W [Jatropha curcas] |
| 9 |
Hb_000175_390 |
0.1202067356 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 10 |
Hb_004374_150 |
0.1208817742 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 11 |
Hb_091384_010 |
0.1230977988 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 12 |
Hb_000261_520 |
0.1259594107 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 13 |
Hb_000088_200 |
0.1263756107 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_073171_080 |
0.1263841647 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_188964_010 |
0.1271906684 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 16 |
Hb_009151_020 |
0.1278767392 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 17 |
Hb_028707_090 |
0.1299624748 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 18 |
Hb_000906_060 |
0.1306687236 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 19 |
Hb_007380_080 |
0.1309002583 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 20 |
Hb_000803_110 |
0.1318955797 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |