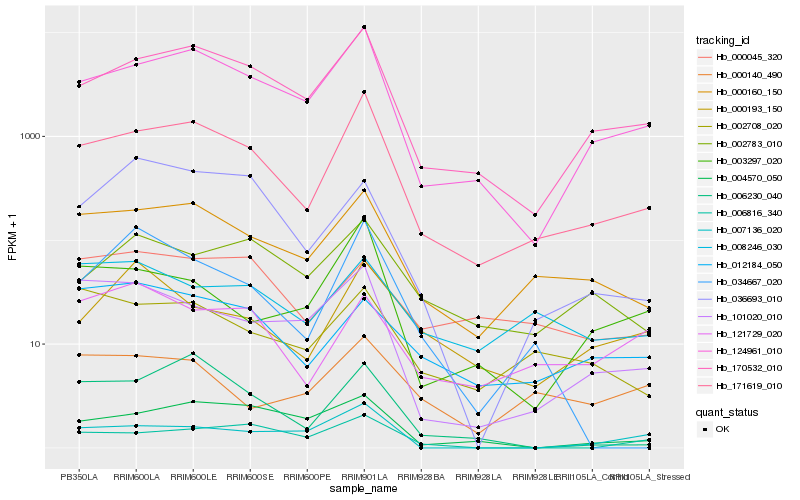
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171619_010 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_124961_010 |
0.1042377136 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 3 |
Hb_170532_010 |
0.1122265664 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 4 |
Hb_000160_150 |
0.1396567803 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 5 |
Hb_003297_020 |
0.1882468812 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 6 |
Hb_101020_010 |
0.1934637581 |
- |
- |
- |
| 7 |
Hb_006230_040 |
0.2034698372 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_004570_050 |
0.2061192123 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_002708_020 |
0.2080759216 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002783_010 |
0.2090236949 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 11 |
Hb_007136_020 |
0.2138282903 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 12 |
Hb_034667_020 |
0.2142751799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006816_340 |
0.2195010532 |
- |
- |
- |
| 14 |
Hb_000193_150 |
0.2217393936 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |
| 15 |
Hb_008246_030 |
0.2249682108 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 16 |
Hb_012184_050 |
0.2284517178 |
- |
- |
hypothetical protein POPTR_0008s01610g [Populus trichocarpa] |
| 17 |
Hb_121729_020 |
0.2293006587 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000140_490 |
0.2348292275 |
- |
- |
BnaC01g32630D [Brassica napus] |
| 19 |
Hb_000045_320 |
0.2368679047 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_036693_010 |
0.2375088143 |
- |
- |
- |