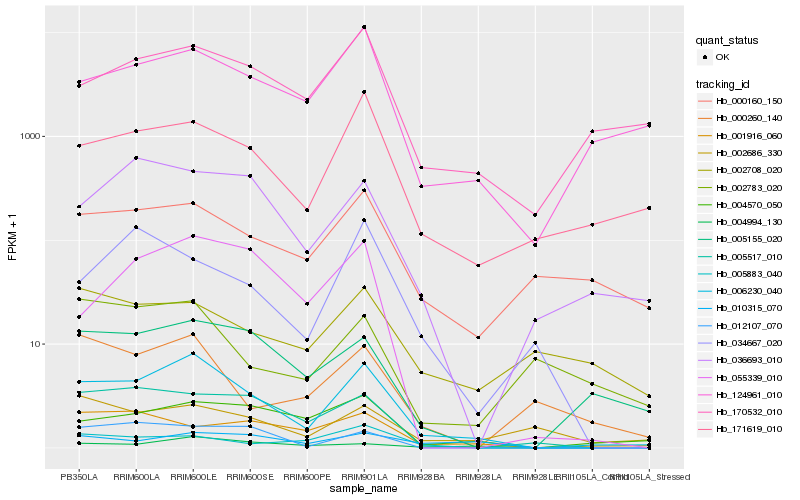
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006230_040 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_171619_010 |
0.2034698372 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_124961_010 |
0.2119822477 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 4 |
Hb_005155_020 |
0.2138629609 |
- |
- |
- |
| 5 |
Hb_170532_010 |
0.2196995707 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 6 |
Hb_004994_130 |
0.2206056197 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 7 |
Hb_004570_050 |
0.2227767907 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_000160_150 |
0.2229114355 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 9 |
Hb_005517_010 |
0.2250040687 |
- |
- |
Chorismate mutase, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_036693_010 |
0.232146561 |
- |
- |
- |
| 11 |
Hb_055339_010 |
0.2366200892 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 12 |
Hb_012107_070 |
0.2385089165 |
- |
- |
- |
| 13 |
Hb_010315_070 |
0.242402682 |
- |
- |
- |
| 14 |
Hb_000260_140 |
0.2469348576 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 15 |
Hb_005883_040 |
0.2482047832 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 16 |
Hb_001916_060 |
0.2573069894 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002686_330 |
0.2626092771 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_034667_020 |
0.2626833507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002708_020 |
0.2636844704 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: uncharacterized protein At5g08430-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002783_020 |
0.2640091962 |
- |
- |
PREDICTED: synaptotagmin-2 isoform X4 [Jatropha curcas] |