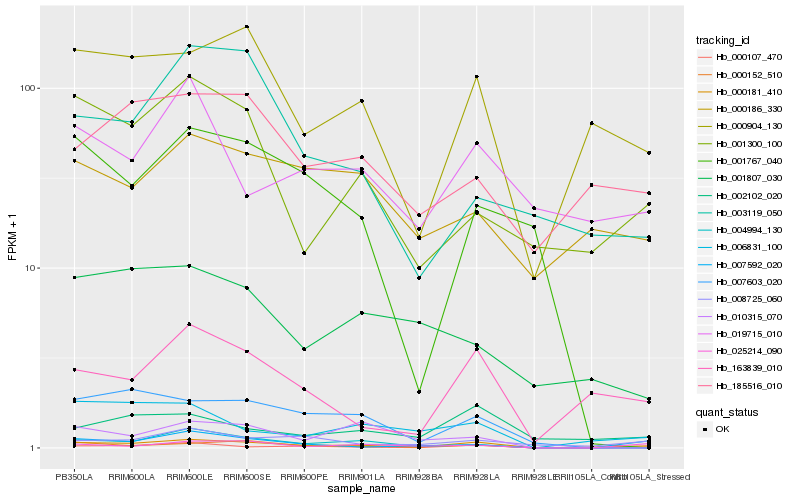
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_410 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000107_470 |
0.2008893765 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 3 |
Hb_007603_020 |
0.2131182516 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 4 |
Hb_004994_130 |
0.2190193462 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 5 |
Hb_000904_130 |
0.2227966808 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 6 |
Hb_000152_510 |
0.230701065 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_163839_010 |
0.230863741 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_006831_100 |
0.2322631695 |
- |
- |
snRNA-activating protein complex subunit, putative [Ricinus communis] |
| 9 |
Hb_010315_070 |
0.2413913035 |
- |
- |
- |
| 10 |
Hb_007592_020 |
0.2428137121 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 11 |
Hb_025214_090 |
0.2503290582 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 12 |
Hb_002102_020 |
0.2520339189 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 13 |
Hb_001300_100 |
0.2533544042 |
- |
- |
- |
| 14 |
Hb_008725_060 |
0.2570374427 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 15 |
Hb_001807_030 |
0.2584072247 |
- |
- |
PREDICTED: CDT1-like protein b [Jatropha curcas] |
| 16 |
Hb_001767_040 |
0.2586319024 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 17 |
Hb_185516_010 |
0.2626468085 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 18 |
Hb_003119_050 |
0.2634457637 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000186_330 |
0.264205156 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 20 |
Hb_019715_010 |
0.2646837991 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |