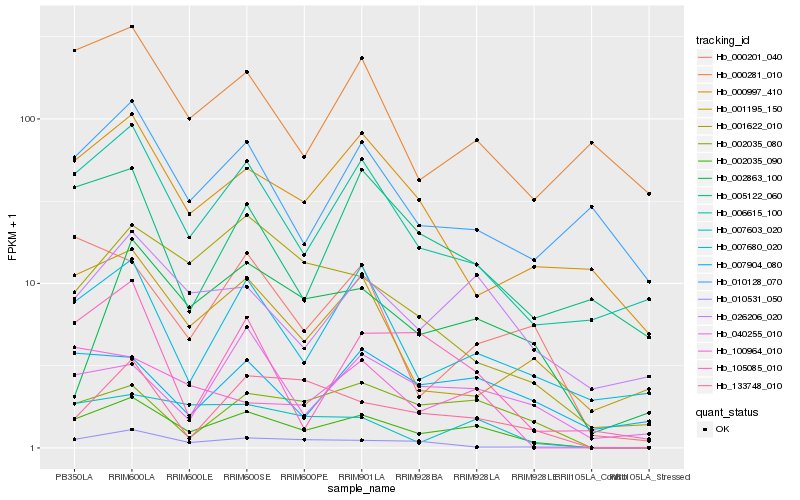
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002035_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_006615_100 |
0.1765425697 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002035_080 |
0.1841968618 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_007603_020 |
0.1864680492 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 5 |
Hb_026206_020 |
0.1920434388 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007680_020 |
0.2010089927 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000997_410 |
0.2115765849 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 8 |
Hb_010531_050 |
0.2117257814 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 9 |
Hb_133748_010 |
0.2172902737 |
- |
- |
- |
| 10 |
Hb_001622_010 |
0.2175068057 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_100964_010 |
0.2189627966 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001195_150 |
0.2199307658 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 13 |
Hb_000201_040 |
0.2212710174 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 14 |
Hb_005122_060 |
0.2251007131 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_040255_010 |
0.2262197334 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 16 |
Hb_000281_010 |
0.2271856246 |
- |
- |
hypothetical protein JCGZ_16194 [Jatropha curcas] |
| 17 |
Hb_010128_070 |
0.2273112254 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002863_100 |
0.2292188583 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 19 |
Hb_105085_010 |
0.2294791185 |
- |
- |
hypothetical protein JCGZ_05865 [Jatropha curcas] |
| 20 |
Hb_007904_080 |
0.2309349322 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |