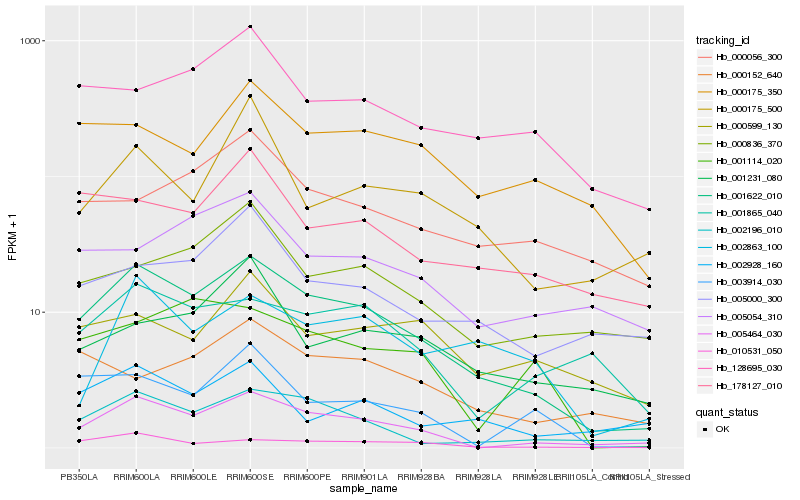
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001622_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_005464_030 |
0.1250993872 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_002196_010 |
0.1599445951 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 4 |
Hb_010531_050 |
0.167123862 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001865_040 |
0.1912316535 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_178127_010 |
0.1932465194 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 7 |
Hb_128695_030 |
0.196971695 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 8 |
Hb_000175_350 |
0.1971227206 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 9 |
Hb_000836_370 |
0.1976648121 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005000_300 |
0.2003442466 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 11 |
Hb_002928_160 |
0.2030968177 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 12 |
Hb_003914_030 |
0.2034035826 |
- |
- |
- |
| 13 |
Hb_001114_020 |
0.2047159246 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 14 |
Hb_005054_310 |
0.206153189 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 15 |
Hb_001231_080 |
0.2092470945 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_002863_100 |
0.2103931387 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 17 |
Hb_000056_300 |
0.2111771401 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000152_640 |
0.2114991292 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 19 |
Hb_000599_130 |
0.2117022559 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_000175_500 |
0.2127002043 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |