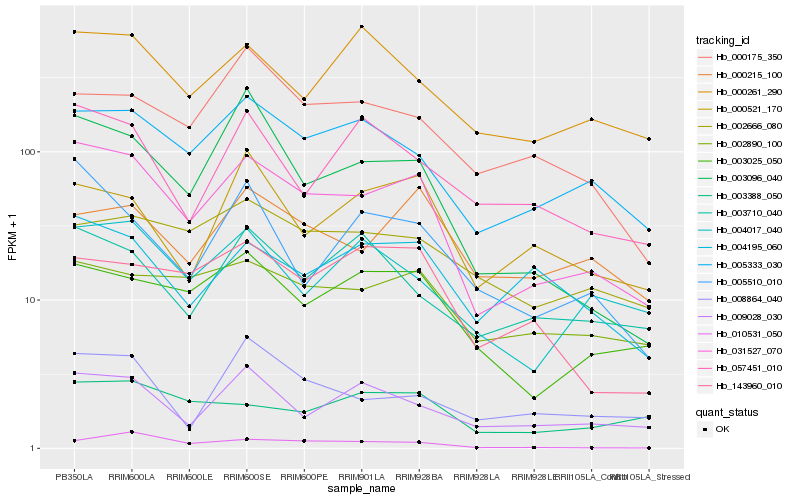
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031527_070 |
0.0 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 2 |
Hb_005333_030 |
0.1352479263 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 3 |
Hb_003025_050 |
0.1498018768 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 4 |
Hb_003096_040 |
0.1531007792 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 5 |
Hb_002890_100 |
0.1584462194 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009028_030 |
0.1619966683 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 7 |
Hb_004195_060 |
0.1622910179 |
- |
- |
PREDICTED: uncharacterized protein LOC105803780 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_004017_040 |
0.1633249787 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 9 |
Hb_005510_010 |
0.1653456547 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010531_050 |
0.1722939646 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 11 |
Hb_143960_010 |
0.1727450695 |
- |
- |
- |
| 12 |
Hb_000261_290 |
0.1733564026 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 13 |
Hb_057451_010 |
0.1736622936 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 14 |
Hb_008864_040 |
0.1749997369 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 15 |
Hb_000175_350 |
0.1751855211 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 16 |
Hb_003710_040 |
0.1757034847 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 17 |
Hb_003388_050 |
0.1759730715 |
- |
- |
- |
| 18 |
Hb_000215_100 |
0.1759985549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002666_080 |
0.1760310423 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000521_170 |
0.1761812364 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |